bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3416_orf1
Length=149
Score E
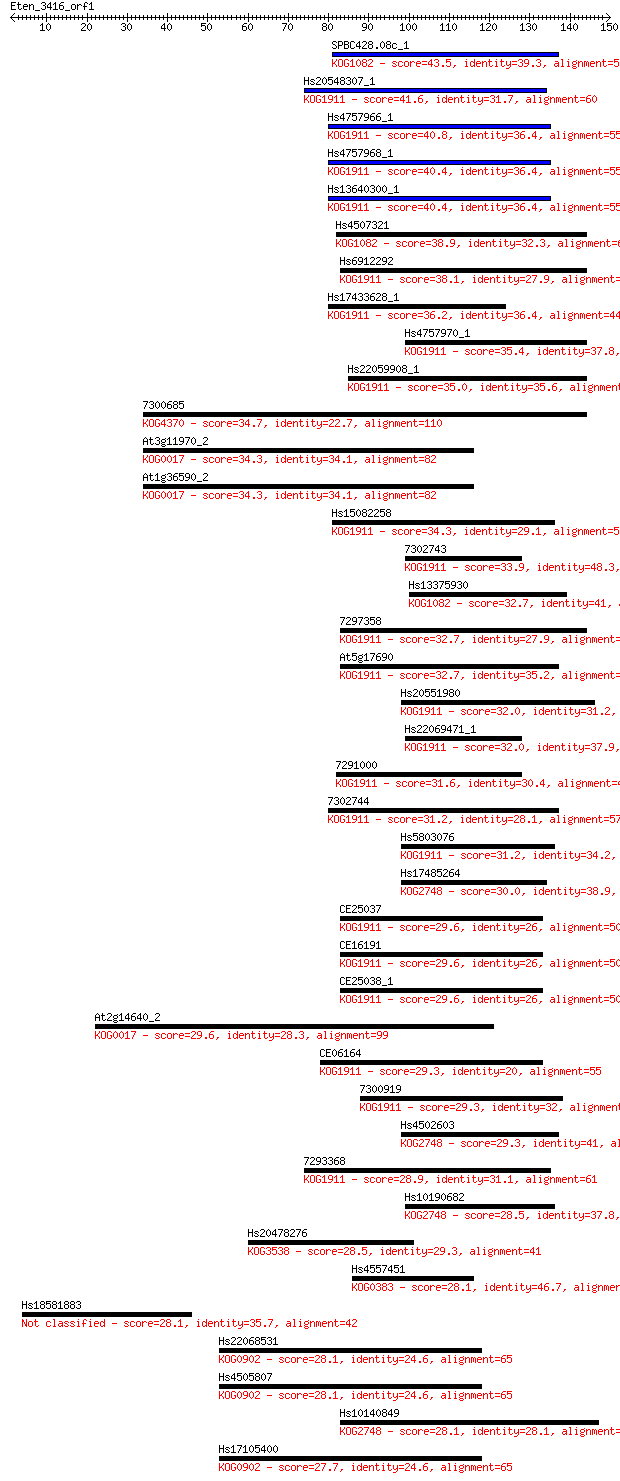
Sequences producing significant alignments: (Bits) Value
SPBC428.08c_1 43.5 1e-04
Hs20548307_1 41.6 5e-04
Hs4757966_1 40.8 0.001
Hs4757968_1 40.4 0.001
Hs13640300_1 40.4 0.001
Hs4507321 38.9 0.003
Hs6912292 38.1 0.006
Hs17433628_1 36.2 0.026
Hs4757970_1 35.4 0.039
Hs22059908_1 35.0 0.049
7300685 34.7 0.075
At3g11970_2 34.3 0.091
At1g36590_2 34.3 0.091
Hs15082258 34.3 0.093
7302743 33.9 0.11
Hs13375930 32.7 0.25
7297358 32.7 0.26
At5g17690 32.7 0.29
Hs20551980 32.0 0.47
Hs22069471_1 32.0 0.48
7291000 31.6 0.56
7302744 31.2 0.73
Hs5803076 31.2 0.82
Hs17485264 30.0 1.6
CE25037 29.6 2.1
CE16191 29.6 2.4
CE25038_1 29.6 2.5
At2g14640_2 29.6 2.5
CE06164 29.3 2.8
7300919 29.3 3.2
Hs4502603 29.3 3.2
7293368 28.9 3.7
Hs10190682 28.5 4.9
Hs20478276 28.5 5.3
Hs4557451 28.1 5.7
Hs18581883 28.1 6.2
Hs22068531 28.1 6.9
Hs4505807 28.1 7.0
Hs10140849 28.1 7.4
Hs17105400 27.7 8.2
> SPBC428.08c_1
Length=126
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 31/58 (53%), Gaps = 2/58 (3%)
Query 81 EEYEANYIMDQCGSGDAAKYLVKCR--GTPEDQATWEPAHHLTGCPTLLRAWRRRQRR 136
EEYE I+D+ + A L + R TWEP +L+GC +L W+RR+RR
Sbjct 6 EEYEVERIVDEKLDRNGAVKLYRIRWLNYSSRSDTWEPPENLSGCSAVLAEWKRRKRR 63
> Hs20548307_1
Length=227
Score = 41.6 bits (96), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 28/60 (46%), Gaps = 0/60 (0%)
Query 74 DSAGSPTEEYEANYIMDQCGSGDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRRR 133
DS + +E I+D G Y V+ +G D TWEP HL C +L +R++
Sbjct 50 DSEEDGEDVFEVEKILDMKTEGGKVLYKVRWKGYTSDDDTWEPEIHLEDCKEVLLEFRKK 109
> Hs4757966_1
Length=226
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 32/57 (56%), Gaps = 3/57 (5%)
Query 80 TEEYEANYIMD--QCGSGDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRRRQ 134
++E+E I+D Q +G+ +YLV+ +G + TWEP HL C + + RRQ
Sbjct 3 SQEFEVEAIVDKRQDKNGNT-QYLVRWKGYDKQDDTWEPEQHLMNCEKCVHDFNRRQ 58
> Hs4757968_1
Length=227
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 32/57 (56%), Gaps = 3/57 (5%)
Query 80 TEEYEANYIMD--QCGSGDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRRRQ 134
++E+E I+D Q +G+ +YLV+ +G + TWEP HL C + + RRQ
Sbjct 3 SQEFEVEAIVDKRQDKNGNT-QYLVRWKGYDKQDDTWEPEQHLMNCEKCVHDFNRRQ 58
> Hs13640300_1
Length=227
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 32/57 (56%), Gaps = 3/57 (5%)
Query 80 TEEYEANYIMD--QCGSGDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRRRQ 134
++E+E I+D Q +G+ +YLV+ +G + TWEP HL C + + RRQ
Sbjct 3 SQEFEVEAIVDKRQDKNGNT-QYLVRWKGYDKQDDTWEPEQHLMNCEKCVHDFNRRQ 58
> Hs4507321
Length=412
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 34/62 (54%), Gaps = 1/62 (1%)
Query 82 EYEANYIMDQCGSGDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRRRQRRSLQAR 141
++E Y+ D + YLVK RG P+ ++TWEP +L C +L+ + + R L R
Sbjct 42 DFEVEYLCDYKKIREQEYYLVKWRGYPDSESTWEPRQNLK-CVRILKQFHKDLERELLRR 100
Query 142 NN 143
++
Sbjct 101 HH 102
> Hs6912292
Length=191
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 17/61 (27%), Positives = 32/61 (52%), Gaps = 1/61 (1%)
Query 83 YEANYIMDQCGSGDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRRRQRRSLQARN 142
Y ++D+ +YL+K +G E+ TWEP +L CP L+ + ++ ++ + N
Sbjct 20 YVVEKVLDRRVVKGQVEYLLKWKGFSEEHNTWEPEKNL-DCPELISEFMKKYKKMKEGEN 78
Query 143 N 143
N
Sbjct 79 N 79
> Hs17433628_1
Length=179
Score = 36.2 bits (82), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 24/45 (53%), Gaps = 1/45 (2%)
Query 80 TEEYEANYIMDQ-CGSGDAAKYLVKCRGTPEDQATWEPAHHLTGC 123
++E+E I+D+ G +YLVK + + TWEP HL C
Sbjct 3 SQEFEVETIVDKRQGKNVNIEYLVKWKAYDKQYDTWEPKQHLMNC 47
> Hs4757970_1
Length=284
Score = 35.4 bits (80), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 24/49 (48%), Gaps = 4/49 (8%)
Query 99 KYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRRR----QRRSLQARNN 143
+YLV+ +G + TWEP HL C + + RR Q+ S R N
Sbjct 78 EYLVRWKGYDSEDDTWEPEQHLVNCEEYIHDFNRRHTEKQKESTLTRTN 126
> Hs22059908_1
Length=233
Score = 35.0 bits (79), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 30/66 (45%), Gaps = 7/66 (10%)
Query 85 ANYIMDQCGSGDAAKYLVK---CRGTPEDQATWEPAHHLTGCPTLLRAWRR----RQRRS 137
A + +CG G++ +Y VK G + TWEP HL C + + R RQ+ S
Sbjct 18 ARVSLRRCGWGNSIQYPVKNPQGEGYNSEDDTWEPEQHLVNCQEYIHDFNRHHTERQKES 77
Query 138 LQARNN 143
R N
Sbjct 78 TLTRTN 83
> 7300685
Length=625
Score = 34.7 bits (78), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 25/111 (22%), Positives = 50/111 (45%), Gaps = 11/111 (9%)
Query 34 RLALPSTCECHNVFHVSQLVPHRPRPPFLPEADAAWPPIRDS-AGSPTEEYEANYIMDQC 92
RL + C C+N+F QL+ ++P + P + D+ TE + + ++ Q
Sbjct 352 RLMVALLCHCNNLFADVQLI------KYVPPLTSTSPKLPDTPEDIQTELRKQDSLLSQI 405
Query 93 GSGDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRRRQRRSLQARNN 143
S A ++ K R ++ WE +T LR + ++Q ++ + +N
Sbjct 406 HSEMNAGFITKKR----EEQLWEVQRIITQLKRKLRTFEKKQEKTAEEVDN 452
> At3g11970_2
Length=958
Score = 34.3 bits (77), Expect = 0.091, Method: Composition-based stats.
Identities = 28/85 (32%), Positives = 40/85 (47%), Gaps = 10/85 (11%)
Query 34 RLALPSTCECHNVFHVSQ---LVPHRPRPPFLPEADAAWPPIRDSAGSPTEEYEANYIMD 90
+LALPS + H VFHVSQ LV + LP ++D E+ +++
Sbjct 867 KLALPSYSQVHPVFHVSQLKVLVGNVSTTVHLPSV------MQDVFEKVPEKVVERKMVN 920
Query 91 QCGSGDAAKYLVKCRGTPEDQATWE 115
+ G K LVK P ++ATWE
Sbjct 921 RQGKA-VTKVLVKWSNEPLEEATWE 944
> At1g36590_2
Length=958
Score = 34.3 bits (77), Expect = 0.091, Method: Composition-based stats.
Identities = 28/85 (32%), Positives = 40/85 (47%), Gaps = 10/85 (11%)
Query 34 RLALPSTCECHNVFHVSQ---LVPHRPRPPFLPEADAAWPPIRDSAGSPTEEYEANYIMD 90
+LALPS + H VFHVSQ LV + LP ++D E+ +++
Sbjct 867 KLALPSYSQVHPVFHVSQLKVLVGNVSTTVHLPSV------MQDVFEKVPEKVVERKMVN 920
Query 91 QCGSGDAAKYLVKCRGTPEDQATWE 115
+ G K LVK P ++ATWE
Sbjct 921 RQGKA-VTKVLVKWSNEPLEEATWE 944
> Hs15082258
Length=183
Score = 34.3 bits (77), Expect = 0.093, Method: Compositional matrix adjust.
Identities = 16/55 (29%), Positives = 28/55 (50%), Gaps = 1/55 (1%)
Query 81 EEYEANYIMDQCGSGDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRRRQR 135
EE+ ++D+ +Y +K +G + TWEP +L CP L+ A+ Q+
Sbjct 28 EEFVVEKVLDRRVVNGKVEYFLKWKGFTDADNTWEPEENLD-CPELIEAFLNSQK 81
> 7302743
Length=84
Score = 33.9 bits (76), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 99 KYLVKCRGTPEDQATWEPAHHLTGCPTLL 127
+YL K G P +Q TWEP +L C TL+
Sbjct 38 QYLTKWEGYPIEQCTWEPLENLGKCMTLI 66
> Hs13375930
Length=350
Score = 32.7 bits (73), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 1/39 (2%)
Query 100 YLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRRRQRRSL 138
YLVK +G P+ TWEP +L CP LL+ + + L
Sbjct 4 YLVKWKGWPDSTNTWEPLQNLK-CPLLLQQFSNDKHNYL 41
> 7297358
Length=206
Score = 32.7 bits (73), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 17/64 (26%), Positives = 34/64 (53%), Gaps = 4/64 (6%)
Query 83 YEANYIMDQCGSGDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAW---RRRQRRSLQ 139
Y I+D+ +Y +K +G PE + TWEP ++L C L++ + R+ + +S
Sbjct 24 YAVEKIIDRRVRKGKVEYYLKWKGYPETENTWEPENNLD-CQDLIQQYEASRKDEEKSAA 82
Query 140 ARNN 143
++ +
Sbjct 83 SKKD 86
> At5g17690
Length=445
Score = 32.7 bits (73), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 29/64 (45%), Gaps = 10/64 (15%)
Query 83 YEANYIMDQCGSGDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAW----------RR 132
YE I + +YL+K RG PE TWEP +L ++ A+ R+
Sbjct 108 YEIEAIRRKRVRKGKVQYLIKWRGWPETANTWEPLENLQSIADVIDAFEGSLKPGKPGRK 167
Query 133 RQRR 136
R+R+
Sbjct 168 RKRK 171
> Hs20551980
Length=125
Score = 32.0 bits (71), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 25/48 (52%), Gaps = 1/48 (2%)
Query 98 AKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRRRQRRSLQARNNLG 145
+Y +K +G + TWEP +L CP L+ A+ Q+ Q ++ G
Sbjct 30 VEYFLKWKGFTDADNTWEPEENL-DCPELIEAFLSSQKAGKQKDDSSG 76
> Hs22069471_1
Length=538
Score = 32.0 bits (71), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 11/29 (37%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 99 KYLVKCRGTPEDQATWEPAHHLTGCPTLL 127
+YL++ +G + TWEP HHL C +
Sbjct 173 EYLIRWKGYGSTEDTWEPEHHLLHCEEFI 201
> 7291000
Length=240
Score = 31.6 bits (70), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 14/46 (30%), Positives = 23/46 (50%), Gaps = 1/46 (2%)
Query 82 EYEANYIMDQCGSGDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLL 127
E+ + D+ +Y +K +G P + TWEP +L CP L+
Sbjct 3 EFSVERVEDKRTVNGRTEYYLKWKGYPRSENTWEPVENLD-CPDLI 47
> 7302744
Length=418
Score = 31.2 bits (69), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 16/57 (28%), Positives = 24/57 (42%), Gaps = 0/57 (0%)
Query 80 TEEYEANYIMDQCGSGDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRRRQRR 136
EEY I+ + + LVK G P + TWEP ++ C L+ + R
Sbjct 21 VEEYVVEKILGKRFVNGRPQVLVKWSGFPNENNTWEPLENVGNCMKLVSDFESEVFR 77
> Hs5803076
Length=185
Score = 31.2 bits (69), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Query 98 AKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRRRQR 135
+YL+K +G ++ TWEP +L CP L+ + + Q+
Sbjct 36 VEYLLKWKGFSDEDNTWEPEENLD-CPDLIAEFLQSQK 72
> Hs17485264
Length=259
Score = 30.0 bits (66), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 21/36 (58%), Gaps = 1/36 (2%)
Query 98 AKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRRR 133
+YLVK +G P +TWEP H+ P L+ A+ +
Sbjct 26 VEYLVKWKGWPPKYSTWEPEEHILD-PRLVMAYEEK 60
> CE25037
Length=301
Score = 29.6 bits (65), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 13/51 (25%), Positives = 27/51 (52%), Gaps = 2/51 (3%)
Query 83 YEANYIMDQ-CGSGDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRR 132
+ ++D+ G ++L++ +G PE ++WEP +L C +L + R
Sbjct 19 FMVEKVLDKRTGKAGRDEFLIQWQGFPESDSSWEPRENLQ-CVEMLDEFER 68
> CE16191
Length=175
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 13/51 (25%), Positives = 27/51 (52%), Gaps = 2/51 (3%)
Query 83 YEANYIMDQ-CGSGDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRR 132
+ ++D+ G ++L++ +G PE ++WEP +L C +L + R
Sbjct 19 FMVEKVLDKRTGKAGRDEFLIQWQGFPESDSSWEPRENLQ-CVEMLDEFER 68
> CE25038_1
Length=162
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 13/51 (25%), Positives = 27/51 (52%), Gaps = 2/51 (3%)
Query 83 YEANYIMDQ-CGSGDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRR 132
+ ++D+ G ++L++ +G PE ++WEP +L C +L + R
Sbjct 19 FMVEKVLDKRTGKAGRDEFLIQWQGFPESDSSWEPRENLQ-CVEMLDEFER 68
> At2g14640_2
Length=492
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 28/105 (26%), Positives = 40/105 (38%), Gaps = 14/105 (13%)
Query 22 FRQLCDRAQSHIRLALPSTCECHNVFHVSQLVPHRPRPPFLPEAD---AAWPPIRDSAG- 77
F+ + RL LP H VFHVS L P++ + + PP+R++
Sbjct 379 FQVASKHGEVAYRLTLPEGTRIHPVFHVSLL------KPWVGDGEPDMGQLPPLRNNGEL 432
Query 78 --SPTEEYEANYIMDQCGSGDAAKYLVKCRGTPEDQATWEPAHHL 120
PT E + A LV+ G + ATWE L
Sbjct 433 KLQPTAVLEVRW--RSQDKKRVADLLVQWEGLHIEDATWEEYDQL 475
> CE06164
Length=184
Score = 29.3 bits (64), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 11/55 (20%), Positives = 30/55 (54%), Gaps = 1/55 (1%)
Query 78 SPTEEYEANYIMDQCGSGDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRR 132
S + + ++++ + ++Y +K +G PE + +WEP +L C +++ + +
Sbjct 32 SSSNVFVVEKVLNKRLTRGGSEYYIKWQGFPESECSWEPIENLQ-CDRMIQEYEK 85
> 7300919
Length=237
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 25/51 (49%), Gaps = 2/51 (3%)
Query 88 IMDQ-CGSGDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRRRQRRS 137
IMD+ S +Y +K RG TWEP + CP L++ + + +S
Sbjct 13 IMDKRITSEGKVEYYIKWRGYTSADNTWEPEEN-CDCPNLIQKFEESRAKS 62
> Hs4502603
Length=558
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query 98 AKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRRRQRR 136
+YLVK RG TWEP ++ P LL A++ R+R+
Sbjct 26 VEYLVKWRGWSPKYNTWEPEENILD-PRLLIAFQNRERQ 63
> 7293368
Length=336
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 32/63 (50%), Gaps = 5/63 (7%)
Query 74 DSAGSPTEEYEANYIMDQCGS--GDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWR 131
D + P +++E I+D + GD K K G +D +WEP+ +L C L+ +
Sbjct 224 DDSIDPEKQWEVEKILDHVATKEGDMFKIRWKKYGPKDD--SWEPSKNL-ACDALIEKFM 280
Query 132 RRQ 134
R+Q
Sbjct 281 RKQ 283
> Hs10190682
Length=389
Score = 28.5 bits (62), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 22/37 (59%), Gaps = 1/37 (2%)
Query 99 KYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRRRQR 135
+YLVK +G + +TWEP ++ LL A+ R+R
Sbjct 27 EYLVKWKGWSQKYSTWEPEENILDA-RLLAAFEERER 62
> Hs20478276
Length=645
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 12/41 (29%), Positives = 19/41 (46%), Gaps = 0/41 (0%)
Query 60 PFLPEADAAWPPIRDSAGSPTEEYEANYIMDQCGSGDAAKY 100
P +PEA+ W + GSP +E + ++ S D Y
Sbjct 144 PGVPEAEVTWFRNKSKLGSPHHLHEGSLLLTNVSSSDQGLY 184
> Hs4557451
Length=1944
Score = 28.1 bits (61), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 19/30 (63%), Gaps = 3/30 (10%)
Query 86 NYIMDQCGSGDAAKYLVKCRGTPEDQATWE 115
N+ +D+ G+ YLVK R P DQ+TWE
Sbjct 638 NHSVDKKGN---YHYLVKWRDLPYDQSTWE 664
> Hs18581883
Length=545
Score = 28.1 bits (61), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 20/42 (47%), Gaps = 0/42 (0%)
Query 4 LDFVGPSAPTLTPPMTKLFRQLCDRAQSHIRLALPSTCECHN 45
+DF+G S PT T TK ++ +AQ I L C N
Sbjct 9 VDFLGLSQPTYTQKKTKTWQMSLRQAQKDISLTPSDQARCSN 50
> Hs22068531
Length=523
Score = 28.1 bits (61), Expect = 6.9, Method: Composition-based stats.
Identities = 16/65 (24%), Positives = 31/65 (47%), Gaps = 3/65 (4%)
Query 53 VPHRPRPPFLPEADAAWPPIRDSAGSPTEEYEANYIMDQCGSGDAAKYLVKCRGTPEDQA 112
+P P L + P++ +A +P Y A + + +CG + K ++CR ED+
Sbjct 243 LPSNPEAIVLDVDYKSGTPMQSAAKAP---YLAKFKVKRCGVSELEKEGLRCRSDSEDEC 299
Query 113 TWEPA 117
+ + A
Sbjct 300 STQEA 304
> Hs4505807
Length=854
Score = 28.1 bits (61), Expect = 7.0, Method: Composition-based stats.
Identities = 16/65 (24%), Positives = 31/65 (47%), Gaps = 3/65 (4%)
Query 53 VPHRPRPPFLPEADAAWPPIRDSAGSPTEEYEANYIMDQCGSGDAAKYLVKCRGTPEDQA 112
+P P L + P++ +A +P Y A + + +CG + K ++CR ED+
Sbjct 528 LPSNPEAIVLDIDYKSGTPMQSAAKAP---YLAKFKVKRCGVSELEKEGLRCRSDSEDEC 584
Query 113 TWEPA 117
+ + A
Sbjct 585 STQEA 589
> Hs10140849
Length=412
Score = 28.1 bits (61), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 31/65 (47%), Gaps = 2/65 (3%)
Query 83 YEANYIMDQCGSGDAAKYLVKCRGTPEDQATWEPAHHLTGCPTLLRAWRRRQR-RSLQAR 141
+ A I+ + +YLVK +G +TWEP ++ L+ A+ +++R R L
Sbjct 11 FAAESIIKRRIRKGRIEYLVKWKGWAIKYSTWEPEENILDS-RLIAAFEQKERERELYGP 69
Query 142 NNLGP 146
GP
Sbjct 70 KKRGP 74
> Hs17105400
Length=2044
Score = 27.7 bits (60), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 16/65 (24%), Positives = 31/65 (47%), Gaps = 3/65 (4%)
Query 53 VPHRPRPPFLPEADAAWPPIRDSAGSPTEEYEANYIMDQCGSGDAAKYLVKCRGTPEDQA 112
+P P L + P++ +A +P Y A + + +CG + K ++CR ED+
Sbjct 1718 LPSNPEAIVLDIDYKSGTPMQSAAKAP---YLAKFKVKRCGVSELEKEGLRCRSDSEDEC 1774
Query 113 TWEPA 117
+ + A
Sbjct 1775 STQEA 1779
Lambda K H
0.320 0.134 0.440
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1858150626
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40