bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3378_orf1
Length=313
Score E
Sequences producing significant alignments: (Bits) Value
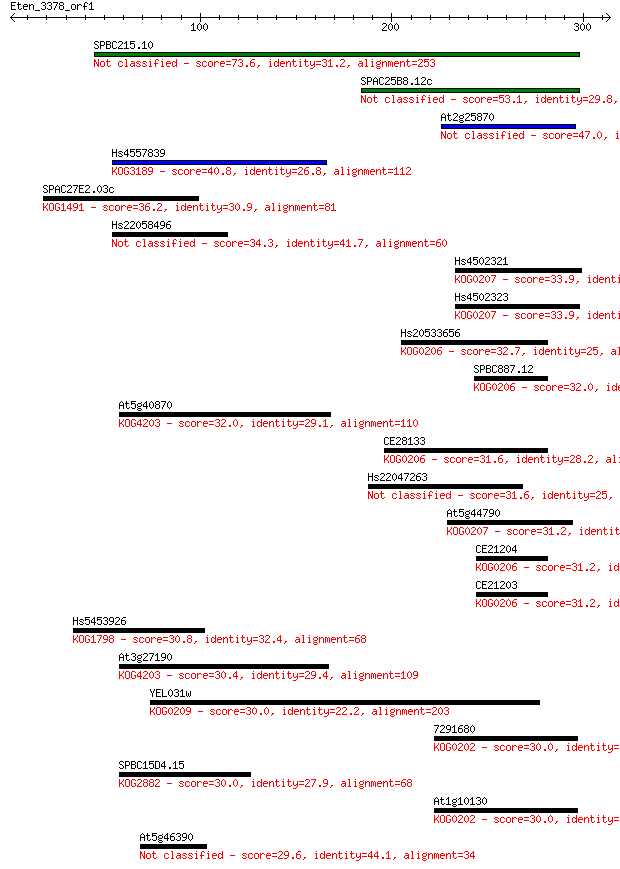
SPBC215.10 73.6 5e-13
SPAC25B8.12c 53.1 8e-07
At2g25870 47.0 6e-05
Hs4557839 40.8 0.004
SPAC27E2.03c 36.2 0.094
Hs22058496 34.3 0.31
Hs4502321 33.9 0.42
Hs4502323 33.9 0.52
Hs20533656 32.7 1.1
SPBC887.12 32.0 1.6
At5g40870 32.0 1.7
CE28133 31.6 2.3
Hs22047263 31.6 2.6
At5g44790 31.2 2.7
CE21204 31.2 3.1
CE21203 31.2 3.2
Hs5453926 30.8 4.3
At3g27190 30.4 4.5
YEL031w 30.0 5.8
7291680 30.0 6.2
SPBC15D4.15 30.0 6.4
At1g10130 30.0 7.0
At5g46390 29.6 8.8
> SPBC215.10
Length=302
Score = 73.6 bits (179), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 79/287 (27%), Positives = 137/287 (47%), Gaps = 41/287 (14%)
Query 45 LPQPSRPIRLIAVDIDGTLT-KNEEFVEKNIEGFKLAKKLGIEVVFV--TGRDPESAKAV 101
LP+P I+LI D+DGTL + + +N+ K ++ ++ FV TGR +
Sbjct 15 LPKPED-IQLIICDVDGTLLGPDHKPHPRNLRALKYLRENYPQLPFVLATGR----QRTS 69
Query 102 VGRHVMEEVGYAGYPGVYVNGAYVVDRDGLVLVDGPLS-----------KDLQERLLQSF 150
VG ++ E +G +P V++NG V D+ G ++ L KD+Q L +
Sbjct 70 VG-NIREALGLHVFPCVHLNGCVVYDK-GEIVACAALKNSLAIDIIDKLKDIQTCALFGY 127
Query 151 D--------------RHGLNTV-VFGETPKGGKGHLREKSDSDSDQYT-LAVFEEPSK-I 193
D +HG+ + + GET K + + D + + VF++ + +
Sbjct 128 DEDYVYQIKQESDKKQHGIKFLRLCGETVKDDANEMLPQLKGPKDIFNKMVVFDDDTNGL 187
Query 194 DDVLPHLVGEFREEIEFTRWAPYAVSVHRMEFSKGSGLTALLQQMH--IPAEEVLALGNG 251
++ L G +E+ T+ P + +KG L +L +++ I E VLA G+G
Sbjct 188 EEAKKRLAGIPSDEVALTQALPQTFEIIPPNDNKGVALKNILSKIYPSISLENVLAFGDG 247
Query 252 VNDLPLFEVAATSVCVRDGELEAVQAADYIT-VNSDEGALLAIVREI 297
ND+ +FE+A SV +R G A++AA I+ V+S EGA+ ++ I
Sbjct 248 ANDVCMFELAGYSVAIRSGMPVALKAAKAISDVSSAEGAVGEVLERI 294
> SPAC25B8.12c
Length=303
Score = 53.1 bits (126), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 34/118 (28%), Positives = 68/118 (57%), Gaps = 5/118 (4%)
Query 184 LAVFEEPSKIDDVLPHLVGEF-REEIEFTRWAPYAVSVHRMEFSKGSGLTALLQQM--HI 240
+AV E+P K+D V+ ++G+F RE+ T+ + + + +KG+ L + + +
Sbjct 173 MAVCEDPDKLD-VVRDILGKFPREKFATTQALEFCIELIPSNSNKGTALQYITSNILPEV 231
Query 241 PAEEVLALGNGVNDLPLFEVAATSVCVRDGELEAVQAADYIT-VNSDEGALLAIVREI 297
E V++ G+G NDL +F +A SV +++G A++ A ++ V ++EGA+ ++ I
Sbjct 232 KNENVISFGDGQNDLSMFAIAGWSVAIKNGMPIAIEKAKAVSRVGNEEGAVGEVLERI 289
> At2g25870
Length=127
Score = 47.0 bits (110), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 44/71 (61%), Gaps = 1/71 (1%)
Query 226 SKGSGLTALLQQMHIPAEEVLALGNGVNDLPLFEVAATSVCVRDGELEAVQAADYITVNS 285
SKG+G+ LL + + +E++A+G+G ND+ + ++A+ V + +G + AD I V++
Sbjct 54 SKGNGVKMLLNHLGVSPDEIMAIGDGENDIEMLQLASLGVALSNGAEKTKAVADVIGVSN 113
Query 286 DE-GALLAIVR 295
D+ G AI R
Sbjct 114 DQDGVADAIYR 124
> Hs4557839
Length=246
Score = 40.8 bits (94), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 30/112 (26%), Positives = 51/112 (45%), Gaps = 5/112 (4%)
Query 54 LIAVDIDGTLTKNEEFVEKNIEGFKLAKKLGIEVVFVTGRDPESAKAVVGRHVMEEVGYA 113
L D+DGTLT + + K ++ F + I++ V G D E + +G V+E+ Y
Sbjct 8 LCLFDVDGTLTAPRQKITKEMDDFLQKLRQKIKIGVVGGSDFEKVQEQLGNDVVEKYDY- 66
Query 114 GYPGVYVNGAYVVDRDGLVLVDGPLSKDLQERLLQSFDRHGLNTVVFGETPK 165
V+ V +DG +L + L E L+Q + L+ + + PK
Sbjct 67 ----VFPENGLVAYKDGKLLCRQNIQSHLGEALIQDLINYCLSYIAKIKLPK 114
> SPAC27E2.03c
Length=392
Score = 36.2 bits (82), Expect = 0.094, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 43/83 (51%), Gaps = 2/83 (2%)
Query 18 LSRAYFSFV-ALFTIFALALAYE-VERAHLPQPSRPIRLIAVDIDGTLTKNEEFVEKNIE 75
L A+ S V A+ I+ + A++ E H+ PIR +++ +D L K+ EFVEK++E
Sbjct 105 LGNAFLSHVRAVDAIYQVVRAFDDAEIIHVEGDVDPIRDLSIIVDELLIKDAEFVEKHLE 164
Query 76 GFKLAKKLGIEVVFVTGRDPESA 98
G + G + + + E A
Sbjct 165 GLRKITSRGANTLEMKAKKEEQA 187
> Hs22058496
Length=116
Score = 34.3 bits (77), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 25/63 (39%), Positives = 38/63 (60%), Gaps = 6/63 (9%)
Query 54 LIAVDIDGTLTKNEEFVEKNIEGF--KLAKKLGIEVVFVTGR-DPESAKAVVGRHVMEEV 110
L +D+DGTLT ++ + K ++ F KL +K+ I VV GR D E A+ +G V+E+
Sbjct 8 LCLLDVDGTLTTLQQKITKEMDDFLQKLRQKIKIGVV---GRLDFEEAQEQLGNAVVEKY 64
Query 111 GYA 113
YA
Sbjct 65 DYA 67
> Hs4502321
Length=1500
Score = 33.9 bits (76), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 36/66 (54%), Gaps = 3/66 (4%)
Query 233 ALLQQMHIPAEEVLALGNGVNDLPLFEVAATSVCVRDGELEAVQAADYITVNSDEGALLA 292
A ++Q+ + V +G+G+ND P +A + + G A++AAD + + +D LL
Sbjct 1284 AKVKQLQEEGKRVAMVGDGINDSPALAMANVGIAIGTGTDVAIEAADVVLIRND---LLD 1340
Query 293 IVREIE 298
+V I+
Sbjct 1341 VVASID 1346
> Hs4502323
Length=1465
Score = 33.9 bits (76), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 35/65 (53%), Gaps = 3/65 (4%)
Query 233 ALLQQMHIPAEEVLALGNGVNDLPLFEVAATSVCVRDGELEAVQAADYITVNSDEGALLA 292
A +Q++ ++V +G+GVND P A V + G A++AAD + + +D LL
Sbjct 1250 AKVQELQNKGKKVAMVGDGVNDSPALAQADMGVAIGTGTDVAIEAADVVLIRND---LLD 1306
Query 293 IVREI 297
+V I
Sbjct 1307 VVASI 1311
> Hs20533656
Length=674
Score = 32.7 bits (73), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 19/77 (24%), Positives = 36/77 (46%), Gaps = 4/77 (5%)
Query 205 REEIEFTRWAPYAVSVHRMEFSKGSGLTALLQQMHIPAEEVLALGNGVNDLPLFEVAATS 264
++ +E T W V K + + + + LA+G+G ND+ + +VA
Sbjct 262 KQFLELTSWCQAVVCCRATPLQKSEVVKLVRSHLQV---MTLAIGDGANDVSMIQVADIG 318
Query 265 VCVRDGE-LEAVQAADY 280
+ V E ++AV A+D+
Sbjct 319 IGVSGQEGMQAVMASDF 335
> SPBC887.12
Length=1258
Score = 32.0 bits (71), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 27/39 (69%), Gaps = 1/39 (2%)
Query 243 EEVLALGNGVNDLPLFEVAATSVCVRDGE-LEAVQAADY 280
E +LA+G+G ND+P+ + A V + E L+AV+++D+
Sbjct 902 EVLLAIGDGANDVPMIQAAHVGVGISGMEGLQAVRSSDF 940
> At5g40870
Length=486
Score = 32.0 bits (71), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 32/123 (26%), Positives = 54/123 (43%), Gaps = 13/123 (10%)
Query 58 DIDGTLTKNEEFVEKNIEGFKLAKKLGIEVVFVTGRDPESAKAVVGRHVMEEVGYAG--- 114
D++ L + +FV+ + F L K +V+ G D A ++ +H+ ++G
Sbjct 211 DVNSVLEQYAKFVKPAFDDFVLPSKKYADVIIPRGGDNHVAVDLITQHIHTKLGQHDLCK 270
Query 115 -YPGVYVNGAYVVDRDGLVLV-DGPLSKD----LQERLLQSFDRHGLNTVVFGE----TP 164
YP VYV + R L+ + +SK +RL++ HGL + F E TP
Sbjct 271 IYPNVYVIQSTFQIRGMHTLIREKDISKHDFVFYSDRLIRLVVEHGLGHLPFTEKQVVTP 330
Query 165 KGG 167
G
Sbjct 331 TGA 333
> CE28133
Length=1139
Score = 31.6 bits (70), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 24/86 (27%), Positives = 43/86 (50%), Gaps = 3/86 (3%)
Query 196 VLPHLVGEFREEIEFTRWAPYAVSVHRMEFSKGSGLTALLQQMHIPAEEVLALGNGVNDL 255
+L L GE R+ +AV RM + + + +++++ VLA+G+G ND+
Sbjct 713 LLHALTGEARKHFGDLALRCHAVVCCRMSPMQKAEVVEMVRKL--AKHVVLAIGDGANDV 770
Query 256 PLFEVAATSVCVRDGE-LEAVQAADY 280
+ + A V + E L+A A+DY
Sbjct 771 AMIQAANVGVGISGEEGLQAASASDY 796
> Hs22047263
Length=313
Score = 31.6 bits (70), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 20/80 (25%), Positives = 34/80 (42%), Gaps = 14/80 (17%)
Query 188 EEPSKIDDVLPHLVGEFREEIEFTRWAPYAVSVHRMEFSKGSGLTALLQQMHIPAEEVLA 247
E+P DV PH G +E W P A+ H + + ++P +
Sbjct 147 EQPRSAGDVTPH--GAIHDE---GGWRPVAIKHH---------CSTAIDHRYVPLVATIK 192
Query 248 LGNGVNDLPLFEVAATSVCV 267
+G+G + LP F ++VC+
Sbjct 193 MGSGHSLLPQFRANNSTVCL 212
> At5g44790
Length=1001
Score = 31.2 bits (69), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 18/66 (27%), Positives = 34/66 (51%), Gaps = 1/66 (1%)
Query 229 SGLTALLQQMHIPAEEVLALGNGVNDLPLFEVAATSVCVRDGELEAVQAADYITV-NSDE 287
+G +++ + V +G+G+ND P A + + G A++AADY+ + N+ E
Sbjct 856 AGKADVIRSLQKDGSTVAMVGDGINDSPALAAADVGMAIGAGTDVAIEAADYVLMRNNLE 915
Query 288 GALLAI 293
+ AI
Sbjct 916 DVITAI 921
> CE21204
Length=1454
Score = 31.2 bits (69), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Query 244 EVLALGNGVNDLPLFEVAATSVCVRDGE-LEAVQAADY 280
VLA+G+G ND+P+ + A + + E L+A A D+
Sbjct 931 NVLAIGDGANDVPMIQAAHVGIGIAGKEGLQAAMACDF 968
> CE21203
Length=1212
Score = 31.2 bits (69), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Query 244 EVLALGNGVNDLPLFEVAATSVCVRDGE-LEAVQAADY 280
VLA+G+G ND+P+ + A + + E L+A A D+
Sbjct 931 NVLAIGDGANDVPMIQAAHVGIGIAGKEGLQAAMACDF 968
> Hs5453926
Length=2286
Score = 30.8 bits (68), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 35/75 (46%), Gaps = 7/75 (9%)
Query 34 LALAYEVERAHLP-----QPSRPIRLIAVDIDGT--LTKNEEFVEKNIEGFKLAKKLGIE 86
+ LA+++E LP + I +I+ IDG L N E V ++IE F+ K E
Sbjct 270 VVLAFDIETTKLPLKFPDAETDQIMMISYMIDGQGYLITNREIVSEDIEDFEFTPKPEYE 329
Query 87 VVFVTGRDPESAKAV 101
F +P+ A +
Sbjct 330 GPFCVFNEPDEAHLI 344
> At3g27190
Length=483
Score = 30.4 bits (67), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 32/122 (26%), Positives = 53/122 (43%), Gaps = 13/122 (10%)
Query 58 DIDGTLTKNEEFVEKNIEGFKLAKKLGIEVVFVTGRDPESAKAVVGRHVMEEVGYAG--- 114
D+D L + +FV+ + F L K +V+ G D A ++ +H+ ++G
Sbjct 211 DVDSVLEQYAKFVKPAFDDFVLPSKKYADVIIPRGGDNHVAVDLIVQHIHTKLGQHDLCK 270
Query 115 -YPGVYVNGAYVVDRDGLVLV-DGPLSKD----LQERLLQSFDRHGLNTVVFGE----TP 164
YP V+V R L+ + +SK +RL++ HGL + F E TP
Sbjct 271 IYPNVFVIETTFQIRGMHTLIREKDISKHDFVFYSDRLIRLVVEHGLGHLPFTEKQVVTP 330
Query 165 KG 166
G
Sbjct 331 TG 332
> YEL031w
Length=1215
Score = 30.0 bits (66), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 45/203 (22%), Positives = 78/203 (38%), Gaps = 40/203 (19%)
Query 74 IEGFKLAKKLGIEVVFVTGRDPESAKAVVGRHVMEEVGYAGYPGVYVNGAYVVDRDGLVL 133
IE K+ + + +TG +P +A HV +EVG + ++DR G
Sbjct 680 IETIKMLNESSHRSIMITGDNPLTA-----VHVAKEVG------IVFGETLILDRAGKSD 728
Query 134 VDGPLSKDLQERLLQSFDRHGLNTVVFGETPKGGKGHLREKSDSDSDQYTLAVFEEPSKI 193
+ L +D++E + FD + F + L ++ D Y L E S++
Sbjct 729 DNQLLFRDVEETVSIPFDP---SKDTFDHS------KLFDRYDIAVTGYALNALEGHSQL 779
Query 194 DDVLPHLVGEFREEIEFTRWAPYAVSVHRMEFSKGSGLTALLQQMHIPAEEVLALGNGVN 253
D+L H W VS + EF LL + + L G+G N
Sbjct 780 RDLLRH------------TWVYARVSPSQKEF--------LLNTLKDMGYQTLMCGDGTN 819
Query 254 DLPLFEVAATSVCVRDGELEAVQ 276
D+ + A + + +G E ++
Sbjct 820 DVGALKQAHVGIALLNGTEEGLK 842
> 7291680
Length=1020
Score = 30.0 bits (66), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 38/76 (50%), Gaps = 5/76 (6%)
Query 222 RMEFSKGSGLTALLQQMHIPAEEVLAL-GNGVNDLPLFEVAATSVCVRDGELEAVQAADY 280
R+E S + LQ M+ E+ A+ G+GVND P + A + + G A AA+
Sbjct 678 RVEPQHKSKIVEFLQSMN----EISAMTGDGVNDAPALKKAEIGIAMGSGTAVAKSAAEM 733
Query 281 ITVNSDEGALLAIVRE 296
+ + + ++++ V E
Sbjct 734 VLADDNFSSIVSAVEE 749
> SPBC15D4.15
Length=298
Score = 30.0 bits (66), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 33/73 (45%), Gaps = 7/73 (9%)
Query 58 DIDGTLTKNEEFVEKNIEGFKLAKKLGIEVVFVTGRDPESAKAVVGRHVMEEVGYAG--- 114
D DG L + + + KL + LG +++FV+ +S + + + + E G A
Sbjct 24 DCDGVLWSGSKPIPGVTDTMKLLRSLGKQIIFVSNNSTKSRETYMNK--INEHGIAAKLE 81
Query 115 --YPGVYVNGAYV 125
YP Y + YV
Sbjct 82 EIYPSAYSSATYV 94
> At1g10130
Length=992
Score = 30.0 bits (66), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 39/76 (51%), Gaps = 5/76 (6%)
Query 222 RMEFSKGSGLTALLQQMHIPAEEVLAL-GNGVNDLPLFEVAATSVCVRDGELEAVQAADY 280
R+E S L LQ+ + EV+A+ G+GVND P + A + + G A A+D
Sbjct 661 RVEPSHKRMLVEALQKQN----EVVAMTGDGVNDAPALKKADIGIAMGSGTAVAKSASDM 716
Query 281 ITVNSDEGALLAIVRE 296
+ + + +++A V E
Sbjct 717 VLADDNFASIVAAVAE 732
> At5g46390
Length=488
Score = 29.6 bits (65), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 69 FVEKNIEGFKLAKKLGIEVVFVTGRDPESAKAVV 102
V+ IE KL G V++ GRDPE+ K VV
Sbjct 349 LVQAGIETAKLFLDEGDTVIYTAGRDPEAQKTVV 382
Lambda K H
0.319 0.138 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7237550830
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40