bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3307_orf6
Length=221
Score E
Sequences producing significant alignments: (Bits) Value
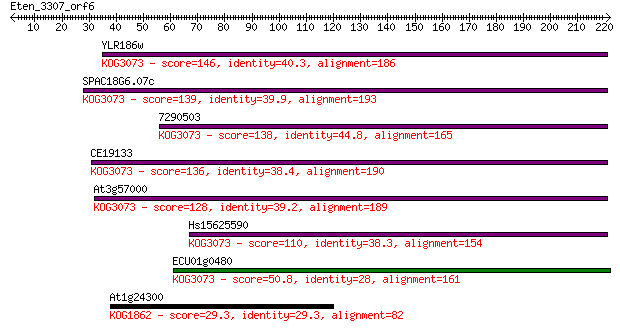
YLR186w 146 3e-35
SPAC18G6.07c 139 4e-33
7290503 138 7e-33
CE19133 136 3e-32
At3g57000 128 1e-29
Hs15625590 110 3e-24
ECU01g0480 50.8 2e-06
At1g24300 29.3 6.8
> YLR186w
Length=252
Score = 146 bits (368), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 75/186 (40%), Positives = 111/186 (59%), Gaps = 1/186 (0%)
Query 35 LLEGFQHRKLLQQENRSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHP 94
LL H+ LL++ R I++ RPDITHQCL+ L +SP+NKAG+L V+I+T G LIEV+P
Sbjct 67 LLNCDDHQGLLKKMGRDISEARPDITHQCLLTLLDSPINKAGKLQVYIQTSRGILIEVNP 126
Query 95 QLRVPPTYDEFRKLMINLLHTRRIRAVEKNVTLMQVTKNDANLFLPVGSRKIALSTQGRP 154
+R+P T+ F LM+ LLH IR+V L++V KN LP RK+ LS
Sbjct 127 TVRIPRTFKRFSGLMVQLLHKLSIRSVNSEEKLLKVIKNPITDHLPTKCRKVTLSFDAPV 186
Query 155 VDLGDFVQQFQDTKNPVLFLVGAVAHANPTTNSELAEECISIAPCGLSAAVCCSSICAEF 214
+ + D++++ D ++ +F VGA+A E +E + ++ LSA+V CS C
Sbjct 187 IRVQDYIEKLDDDESICVF-VGAMARGKDNFADEYVDEKVGLSNYPLSASVACSKFCHGA 245
Query 215 ENLWNI 220
E+ WNI
Sbjct 246 EDAWNI 251
> SPAC18G6.07c
Length=359
Score = 139 bits (351), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 77/193 (39%), Positives = 109/193 (56%), Gaps = 1/193 (0%)
Query 28 GREGSLQLLEGFQHRKLLQQENRSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSG 87
++ QLL H+ +L++ NR+IA RPDITHQCL+ L +SPLNKAGRL V+I T
Sbjct 167 AKDAKYQLLNCDDHQGILKKLNRNIAQARPDITHQCLLTLLDSPLNKAGRLQVYIHTAKK 226
Query 88 QLIEVHPQLRVPPTYDEFRKLMINLLHTRRIRAVEKNVTLMQVTKNDANLFLPVGSRKIA 147
LIEV+P +R+P T+ F LM+ LLH IR+V N L++V KN +LP RK
Sbjct 227 VLIEVNPSVRIPRTFKRFSGLMVQLLHKLSIRSVNGNEKLLKVIKNPVTDYLPPNCRKAT 286
Query 148 LSTQGRPVDLGDFVQQFQDTKNPVLFLVGAVAHANPTTNSELAEECISIAPCGLSAAVCC 207
LS V +++ Q ++ V +GA+AH + +E ISI+ LSA++ C
Sbjct 287 LSFDAPTVPPRKYLETLQPNQS-VCIAIGAMAHGPDDFSDGWVDEKISISDYPLSASIAC 345
Query 208 SSICAEFENLWNI 220
S E+ I
Sbjct 346 SKFLHSMEDFLGI 358
> 7290503
Length=233
Score = 138 bits (348), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 74/170 (43%), Positives = 104/170 (61%), Gaps = 7/170 (4%)
Query 56 RPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHPQLRVPPTYDEFRKLMINLLHT 115
RPDITHQCL+ L +SPLN+AG L VF+RT+ LIE++PQ R+P T+ F LM+ LLH
Sbjct 65 RPDITHQCLLMLFDSPLNRAGLLQVFVRTEHNVLIEINPQTRIPRTFKRFAGLMVQLLHK 124
Query 116 RRIRAVEKNVTLMQVTKNDANLFLPVGSRKIALSTQGRPV-DLGDFVQQFQDTK----NP 170
+IRA + + LM V KN +PVG +K A+S G+ + + D V +T P
Sbjct 125 FQIRANDSSRRLMSVIKNPITDHVPVGCKKYAMSFSGKLLPNCRDLVPHGDETSASYDEP 184
Query 171 VLFLVGAVAHANPTTNSELAEECISIAPCGLSAAVCCSSICAEFENLWNI 220
V+ ++GA AH ++ EE SI+ LSAA+ CS IC+ FE +W +
Sbjct 185 VVIVIGAFAHG--VLKTDYTEELFSISNYPLSAAIACSKICSAFEEVWGV 232
> CE19133
Length=231
Score = 136 bits (343), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 73/190 (38%), Positives = 108/190 (56%), Gaps = 3/190 (1%)
Query 31 GSLQLLEGFQHRKLLQQENRSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLI 90
G +L +H L+++ + AD RPDI HQCL+ L +SPLN+AG+L VF RT L+
Sbjct 44 GEYAILSSDKHANFLRKQKKDPADYRPDILHQCLLNLLDSPLNRAGKLRVFFRTSKNVLV 103
Query 91 EVHPQLRVPPTYDEFRKLMINLLHTRRIRAVEKNVTLMQVTKNDANLFLPVGSRKIALST 150
+V PQ R+P T+D F LM+ LLH IRA E LM V KN + LPVGSRK+ +S
Sbjct 104 DVSPQCRIPRTFDRFCGLMVQLLHKLSIRAAETTQKLMSVVKNPVSNHLPVGSRKMLMSF 163
Query 151 QGRPVDLGDFVQQFQDTKNPVLFLVGAVAHANPTTNSELAEECISIAPCGLSAAVCCSSI 210
+ + + + +T P++ ++G +A + +E IS P LSAA+ C+ +
Sbjct 164 NVPELTMANKLVA-PETDEPLVLIIGGIARGKIVVDYNDSETKISNYP--LSAALTCAKV 220
Query 211 CAEFENLWNI 220
+ E +W I
Sbjct 221 TSGLEEIWGI 230
> At3g57000
Length=298
Score = 128 bits (321), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 74/192 (38%), Positives = 105/192 (54%), Gaps = 5/192 (2%)
Query 32 SLQLLEGFQHRKLLQQENRSIADVRPDITHQCLIALQESPLNKAGRL-CVFIRTQSGQLI 90
+ QLL H L++ NR+ AD RPDITHQ L+ + +SP+NKAGRL V++RT+ G L
Sbjct 108 TYQLLNSDDHANFLKKNNRNPADYRPDITHQALLMILDSPVNKAGRLKAVYVRTEKGVLF 167
Query 91 EVHPQLRVPPTYDEFRKLMINLLHTRRIRAVEKNVTLMQVTKND-ANLFLPVGSRKIALS 149
EV P +R+P T+ F +M+ LL I AV L++ KN LPV S +I S
Sbjct 168 EVKPHVRIPRTFKRFAGIMLQLLQKLSITAVNSREKLLRCVKNPIEEHHLPVNSHRIGFS 227
Query 150 -TQGRPVDLGDFVQQFQDTKNPVLFLVGAVAHANPTTNSELAEECISIAPCGLSAAVCCS 208
+ + V++ + D +F+VGA+AH N +E +S++ LSAA C S
Sbjct 228 HSSEKLVNMQKHLATVCDDDRDTVFVVGAMAHGKIDCN--YIDEFVSVSEYPLSAAYCIS 285
Query 209 SICAEFENLWNI 220
IC WNI
Sbjct 286 RICEALATNWNI 297
> Hs15625590
Length=151
Score = 110 bits (275), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 59/154 (38%), Positives = 90/154 (58%), Gaps = 5/154 (3%)
Query 67 LQESPLNKAGRLCVFIRTQSGQLIEVHPQLRVPPTYDEFRKLMINLLHTRRIRAVEKNVT 126
L +SPLN+AG L V+I TQ LIEV+PQ R+P T+D F LM+ LLH +RA +
Sbjct 2 LMDSPLNRAGLLQVYIHTQKNVLIEVNPQTRIPRTFDRFCGLMVQLLHKLSVRAADGPQK 61
Query 127 LMQVTKNDANLFLPVGSRKIALSTQGRPVDLGDFVQQFQDTKNPVLFLVGAVAHANPTTN 186
L++V KN + PVG K+ S + + V++ + +P++F+VGA AH +
Sbjct 62 LLKVIKNPVSDHFPVGCMKVGTSFS---IPVVSDVRELVPSSDPIVFVVGAFAHGK--VS 116
Query 187 SELAEECISIAPCGLSAAVCCSSICAEFENLWNI 220
E E+ +SI+ LSAA+ C+ + FE +W +
Sbjct 117 VEYTEKMVSISNYPLSAALTCAKLTTAFEEVWGV 150
> ECU01g0480
Length=183
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 45/162 (27%), Positives = 69/162 (42%), Gaps = 10/162 (6%)
Query 61 HQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHPQLRVPPTYDEFRKLMINLLHTRRIRA 120
H+ L L +SPLNK+ ++ ++I T L+E+ L VP + M LL I++
Sbjct 31 HRALRLLMDSPLNKSKKMKIYIHTARNALVELSYLLEVPENPADLSDAMSYLLKRMVIKS 90
Query 121 VEKNVTLMQVTKNDANLFLPVGSRKIALSTQGRPVDLGDFVQQFQDTKNPVLFL-VGAVA 179
+ L +V KN LP S KI LS +G + D + + V F+ VG
Sbjct 91 SD-GTMLGKVVKNPVTNHLPPNSTKIRLSPRGCKMSSKDLESSLE--RGFVFFIDVGREE 147
Query 180 HANPTTNSELAEECISIAPCGLSAAVCCSSICAEFENLWNIY 221
+ ++ LS CC+ I FE L I+
Sbjct 148 EREEAEFD------MKLSDFKLSPESCCAKITNMFEELLQIF 183
> At1g24300
Length=1417
Score = 29.3 bits (64), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 39/82 (47%), Gaps = 8/82 (9%)
Query 38 GFQHRKLLQQENRSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHPQLR 97
G +H LL+ E R I + P + L L+ P + R R ++ H Q +
Sbjct 892 GVRHNTLLE-EQRHIDPLWPSDHNDQL--LRSHPGIQRSRSSTGFRQ-----LDFHQQQQ 943
Query 98 VPPTYDEFRKLMINLLHTRRIR 119
PP D+F +L NLL+ +++R
Sbjct 944 RPPFEDQFGQLERNLLYQQQLR 965
Lambda K H
0.321 0.135 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4183546302
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40