bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
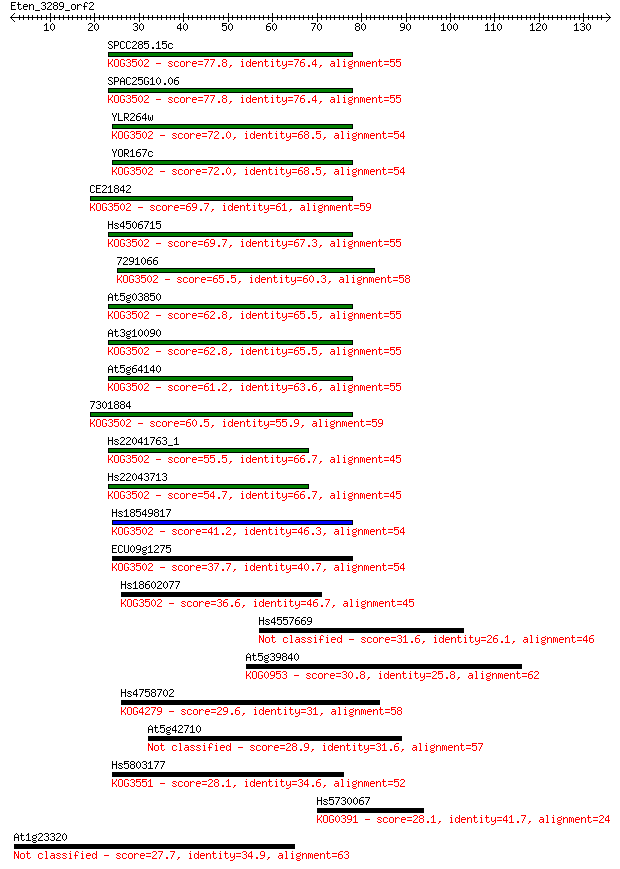
Query= Eten_3289_orf2
Length=135
Score E
Sequences producing significant alignments: (Bits) Value
SPCC285.15c 77.8 5e-15
SPAC25G10.06 77.8 5e-15
YLR264w 72.0 3e-13
YOR167c 72.0 3e-13
CE21842 69.7 2e-12
Hs4506715 69.7 2e-12
7291066 65.5 3e-11
At5g03850 62.8 2e-10
At3g10090 62.8 2e-10
At5g64140 61.2 5e-10
7301884 60.5 9e-10
Hs22041763_1 55.5 3e-08
Hs22043713 54.7 5e-08
Hs18549817 41.2 6e-04
ECU09g1275 37.7 0.005
Hs18602077 36.6 0.013
Hs4557669 31.6 0.41
At5g39840 30.8 0.72
Hs4758702 29.6 1.8
At5g42710 28.9 2.6
Hs5803177 28.1 4.7
Hs5730067 28.1 5.4
At1g23320 27.7 6.5
> SPCC285.15c
Length=68
Score = 77.8 bits (190), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 42/55 (76%), Positives = 45/55 (81%), Gaps = 4/55 (7%)
Query 23 KLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILVLLQ 77
KLAKV KVLGRTGSRGGV QVRV FM + S RS+IRNVKGPVRE DILVLL+
Sbjct 9 KLAKVIKVLGRTGSRGGVTQVRVEFMDDTS----RSIIRNVKGPVREDDILVLLE 59
> SPAC25G10.06
Length=68
Score = 77.8 bits (190), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 42/55 (76%), Positives = 45/55 (81%), Gaps = 4/55 (7%)
Query 23 KLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILVLLQ 77
KLAKV KVLGRTGSRGGV QVRV FM + S RS+IRNVKGPVRE DILVLL+
Sbjct 9 KLAKVIKVLGRTGSRGGVTQVRVEFMDDTS----RSIIRNVKGPVREDDILVLLE 59
> YLR264w
Length=67
Score = 72.0 bits (175), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 37/54 (68%), Positives = 44/54 (81%), Gaps = 4/54 (7%)
Query 24 LAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILVLLQ 77
LAKV KVLGRTGSRGGV QVRV F+ + S R+++RNVKGPVRE DILVL++
Sbjct 9 LAKVIKVLGRTGSRGGVTQVRVEFLEDTS----RTIVRNVKGPVRENDILVLME 58
> YOR167c
Length=67
Score = 72.0 bits (175), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 37/54 (68%), Positives = 44/54 (81%), Gaps = 4/54 (7%)
Query 24 LAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILVLLQ 77
LAKV KVLGRTGSRGGV QVRV F+ + S R+++RNVKGPVRE DILVL++
Sbjct 9 LAKVIKVLGRTGSRGGVTQVRVEFLEDTS----RTIVRNVKGPVRENDILVLME 58
> CE21842
Length=65
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 36/59 (61%), Positives = 46/59 (77%), Gaps = 3/59 (5%)
Query 19 MDFSKLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILVLLQ 77
MD LA+V KV+GRTGS+G QVRV F+ ++++ RS+IRNVKGPVREGDIL LL+
Sbjct 1 MDKLTLARVTKVIGRTGSQGQCTQVRVEFINDQNN---RSIIRNVKGPVREGDILTLLE 56
> Hs4506715
Length=69
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/55 (67%), Positives = 43/55 (78%), Gaps = 4/55 (7%)
Query 23 KLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILVLLQ 77
KLA+V KVLGRTGS+G QVRV FM + S RS+IRNVKGPVREGD+L LL+
Sbjct 10 KLARVTKVLGRTGSQGQCTQVRVEFMDDTS----RSIIRNVKGPVREGDVLTLLE 60
> 7291066
Length=103
Score = 65.5 bits (158), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 35/58 (60%), Positives = 41/58 (70%), Gaps = 4/58 (6%)
Query 25 AKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILVLLQRRQLL 82
A+V KVLGRTGS+G QV+V F+ E Q R +IRNVKGPVREGDIL LL+ L
Sbjct 8 ARVMKVLGRTGSQGQCTQVKVEFLGE----QNRQIIRNVKGPVREGDILTLLESEHLF 61
> At5g03850
Length=64
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 36/55 (65%), Positives = 41/55 (74%), Gaps = 5/55 (9%)
Query 23 KLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILVLLQ 77
K A V KV+GRTGSRG V QVRV F +SD R ++RNVKGPVREGDIL LL+
Sbjct 6 KHAVVVKVMGRTGSRGQVTQVRVKFT--DSD---RYIMRNVKGPVREGDILTLLE 55
> At3g10090
Length=64
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 36/55 (65%), Positives = 41/55 (74%), Gaps = 5/55 (9%)
Query 23 KLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILVLLQ 77
K A V KV+GRTGSRG V QVRV F +SD R ++RNVKGPVREGDIL LL+
Sbjct 6 KHAVVVKVMGRTGSRGQVTQVRVKFT--DSD---RYIMRNVKGPVREGDILTLLE 55
> At5g64140
Length=64
Score = 61.2 bits (147), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 35/55 (63%), Positives = 41/55 (74%), Gaps = 5/55 (9%)
Query 23 KLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILVLLQ 77
K A V KV+GRTGSRG V QVRV F +SD R ++RNVKGPVREGD+L LL+
Sbjct 6 KHAVVVKVMGRTGSRGQVTQVRVKFT--DSD---RFIMRNVKGPVREGDVLTLLE 55
> 7301884
Length=64
Score = 60.5 bits (145), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 33/59 (55%), Positives = 43/59 (72%), Gaps = 4/59 (6%)
Query 19 MDFSKLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILVLLQ 77
MD + A+V ++LGRTGS+G QVRV F+ ++S R +IRNVKGPVR GDIL LL+
Sbjct 1 MDKPQYARVVEILGRTGSQGQCTQVRVEFLGDQS----RQIIRNVKGPVRVGDILSLLE 55
> Hs22041763_1
Length=188
Score = 55.5 bits (132), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 30/45 (66%), Positives = 34/45 (75%), Gaps = 4/45 (8%)
Query 23 KLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPV 67
KLA+V KVLGRTGS+G QVRV FM D+ RS+IRNVKGPV
Sbjct 10 KLARVTKVLGRTGSQGQCTQVRVKFM----DVTSRSIIRNVKGPV 50
> Hs22043713
Length=106
Score = 54.7 bits (130), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 30/45 (66%), Positives = 34/45 (75%), Gaps = 4/45 (8%)
Query 23 KLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPV 67
KLA+V KVLGRTGS+G QVRV FM D+ RS+IRNVKGPV
Sbjct 10 KLARVTKVLGRTGSQGQCTQVRVKFM----DVTSRSIIRNVKGPV 50
> Hs18549817
Length=96
Score = 41.2 bits (95), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 25/54 (46%), Positives = 34/54 (62%), Gaps = 7/54 (12%)
Query 24 LAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILVLLQ 77
LA+V KVLGRT S+G Q FM + S S+I +VK P+R+G +L LL+
Sbjct 41 LARVTKVLGRTSSQG---QCMREFMEDTS----HSIIHSVKDPMRKGYVLTLLE 87
> ECU09g1275
Length=65
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 22/54 (40%), Positives = 32/54 (59%), Gaps = 5/54 (9%)
Query 24 LAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILVLLQ 77
+V VLGRTG G + QV++ M + R++ R VKGPV GDI+ +L+
Sbjct 8 FGEVTHVLGRTGGSGLLTQVKMELMHNK-----RTIQRAVKGPVAVGDIIEILE 56
> Hs18602077
Length=97
Score = 36.6 bits (83), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 21/45 (46%), Positives = 26/45 (57%), Gaps = 4/45 (8%)
Query 26 KVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREG 70
KV +V GRT S+G Q+R M D RS+I N KGPV +G
Sbjct 57 KVTEVPGRTSSQGQGTQLRTKLM----DYTSRSIIHNAKGPVPQG 97
> Hs4557669
Length=825
Score = 31.6 bits (70), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 12/46 (26%), Positives = 21/46 (45%), Gaps = 0/46 (0%)
Query 57 RSLIRNVKGPVREGDILVLLQRRQLLQRRRRQRGRAAARCCSWTGC 102
+ + P R + +++Q Q Q +R RG+ A+C W C
Sbjct 260 KEWWDQIPNPARSRLVAIIIQDAQGSQWEKRSRGQEPAKCPHWKNC 305
> At5g39840
Length=776
Score = 30.8 bits (68), Expect = 0.72, Method: Composition-based stats.
Identities = 16/62 (25%), Positives = 29/62 (46%), Gaps = 1/62 (1%)
Query 54 LQGRSLIRNVKGPVREGDILVLLQRRQLLQRRRRQRGRAAARCCSWTGCRPVGQQRQAVL 113
++ ++L+ +K V+ GD +V RR++ + + RCC G P +RQ
Sbjct 423 VEAKTLLGELKN-VKSGDCVVAFSRREIFEVKMAIEKHTNHRCCVIYGALPPETRRQQAK 481
Query 114 LL 115
L
Sbjct 482 LF 483
> Hs4758702
Length=1011
Score = 29.6 bits (65), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 32/58 (55%), Gaps = 4/58 (6%)
Query 26 KVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILVLLQRRQLLQ 83
+ + LG + S+GG ++ FM E SL+R+V GP+++ + + RQ+LQ
Sbjct 427 NIVRYLG-SASQGGYLKI---FMEEVPGGSLSSLLRSVWGPLKDNESTISFYTRQILQ 480
> At5g42710
Length=807
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 31/57 (54%), Gaps = 4/57 (7%)
Query 32 GRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILVLLQRRQLLQRRRRQ 88
GR + G QQ + SD+ +++ + KG + G L LL++RQ +RR+R+
Sbjct 577 GRMFTNGMDQQAPIPKSDGNSDILSKTVYKETKGEIEAG--LPLLEKRQ--ERRKRE 629
> Hs5803177
Length=540
Score = 28.1 bits (61), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 32/54 (59%), Gaps = 2/54 (3%)
Query 24 LAKVEKVLGRTGSRGGVQQVR-VTFMTEESDLQ-GRSLIRNVKGPVREGDILVL 75
LA++ +LG T + GG ++V+ + ++ E++ L GR R V V E D+L+
Sbjct 307 LAELNAMLGATSTAGGSKEVKHIAWLAEQAKLDGGRQQWRPVLMAVTEKDLLLY 360
> Hs5730067
Length=2971
Score = 28.1 bits (61), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 10/24 (41%), Positives = 20/24 (83%), Gaps = 0/24 (0%)
Query 70 GDILVLLQRRQLLQRRRRQRGRAA 93
G ++ +++ ++L+QRRR+QRG A+
Sbjct 2493 GRLVTVVEEKELVQRRRQQRGAAS 2516
> At1g23320
Length=388
Score = 27.7 bits (60), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 35/69 (50%), Gaps = 6/69 (8%)
Query 2 QTPNPSPKPQTLAPNPKMDFSKLAKVEKVLGRTGSRGGVQQ--VRVTFMTEESD----LQ 55
+T + SP L + D L K +KVL R G R G + VRV+ ++ + D LQ
Sbjct 316 KTLSTSPAFAWLGYKEERDLGSLLKEKKVLTRGGDRCGCNKRYVRVSMLSRDDDFDVSLQ 375
Query 56 GRSLIRNVK 64
+ I+++K
Sbjct 376 RLATIKDLK 384
Lambda K H
0.322 0.135 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1388795348
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40