bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3268_orf1
Length=137
Score E
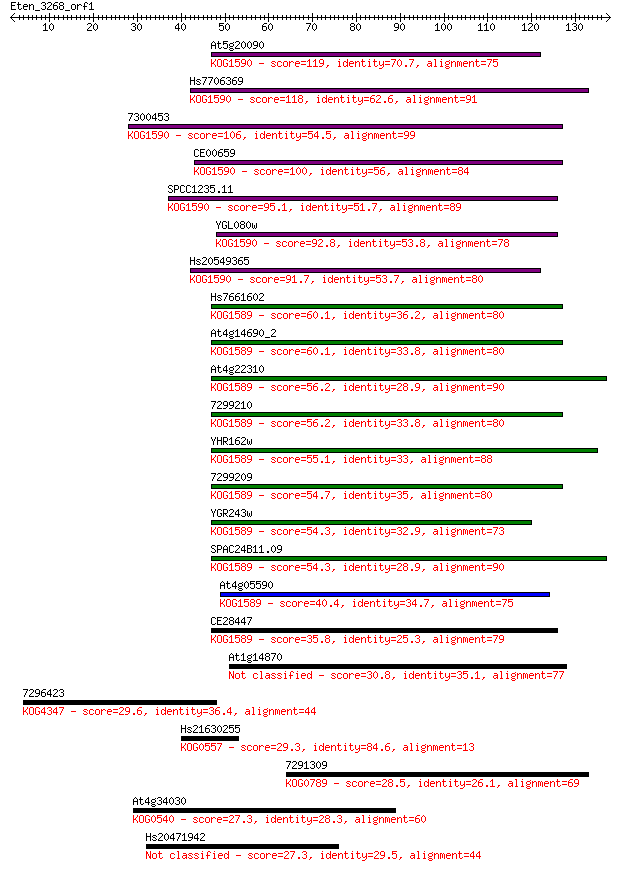
Sequences producing significant alignments: (Bits) Value
At5g20090 119 1e-27
Hs7706369 118 3e-27
7300453 106 1e-23
CE00659 100 6e-22
SPCC1235.11 95.1 3e-20
YGL080w 92.8 1e-19
Hs20549365 91.7 4e-19
Hs7661602 60.1 1e-09
At4g14690_2 60.1 1e-09
At4g22310 56.2 2e-08
7299210 56.2 2e-08
YHR162w 55.1 4e-08
7299209 54.7 5e-08
YGR243w 54.3 6e-08
SPAC24B11.09 54.3 7e-08
At4g05590 40.4 0.001
CE28447 35.8 0.027
At1g14870 30.8 0.69
7296423 29.6 1.8
Hs21630255 29.3 2.2
7291309 28.5 3.8
At4g34030 27.3 9.4
Hs20471942 27.3 9.5
> At5g20090
Length=110
Score = 119 bits (299), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 53/75 (70%), Positives = 59/75 (78%), Gaps = 0/75 (0%)
Query 47 RTTHFWGPVANWGFVIAGLSDMRKSPECISERMTGVLCIYSLLFMRFATMVQPRNSLLFA 106
+TTHFWGP+ANWGFV AGL DM+K PE IS M+ +C+YS LFMRFA MVQPRN LL A
Sbjct 17 KTTHFWGPIANWGFVAAGLVDMQKPPEMISGNMSSAMCVYSALFMRFAWMVQPRNYLLLA 76
Query 107 CHACNEAVQLTQLYR 121
CHA NE VQL QL R
Sbjct 77 CHASNETVQLYQLSR 91
> Hs7706369
Length=109
Score = 118 bits (296), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 57/91 (62%), Positives = 63/91 (69%), Gaps = 0/91 (0%)
Query 42 LRTYGRTTHFWGPVANWGFVIAGLSDMRKSPECISERMTGVLCIYSLLFMRFATMVQPRN 101
R Y +THFWGPVANWG IA ++DM+KSPE IS RMT LC YSL FMRFA VQPRN
Sbjct 18 FRDYLMSTHFWGPVANWGLPIAAINDMKKSPEIISGRMTFALCCYSLTFMRFAYKVQPRN 77
Query 102 SLLFACHACNEAVQLTQLYRKLSYNATKANS 132
LLFACHA NE QL Q R + + TK S
Sbjct 78 WLLFACHATNEVAQLIQGGRLIKHEMTKTAS 108
> 7300453
Length=107
Score = 106 bits (264), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 54/99 (54%), Positives = 64/99 (64%), Gaps = 7/99 (7%)
Query 28 ATMASRGWSEFVTYLRTYGRTTHFWGPVANWGFVIAGLSDMRKSPECISERMTGVLCIYS 87
+T AS+ W R Y +THFWGPVANWG +A L+D +KSP+ IS +MT L +YS
Sbjct 8 STTASKEW-------RDYFMSTHFWGPVANWGIPVAALADTQKSPKFISGKMTLALTLYS 60
Query 88 LLFMRFATMVQPRNSLLFACHACNEAVQLTQLYRKLSYN 126
+FMRFA VQPRN LLFACHA N Q Q R L YN
Sbjct 61 CIFMRFAYKVQPRNWLLFACHATNATAQSIQGLRFLHYN 99
> CE00659
Length=160
Score = 100 bits (250), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 47/84 (55%), Positives = 59/84 (70%), Gaps = 0/84 (0%)
Query 43 RTYGRTTHFWGPVANWGFVIAGLSDMRKSPECISERMTGVLCIYSLLFMRFATMVQPRNS 102
+ Y +THFWGPVANWG +A L D++K+P+ IS MT L IYS +FMRFA VQPRN
Sbjct 21 KHYFLSTHFWGPVANWGLPLAALGDLKKNPDMISGPMTSALLIYSSVFMRFAWHVQPRNL 80
Query 103 LLFACHACNEAVQLTQLYRKLSYN 126
LLFACH N + Q QL R +++N
Sbjct 81 LLFACHFANFSAQGAQLGRFVNHN 104
> SPCC1235.11
Length=141
Score = 95.1 bits (235), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 46/94 (48%), Positives = 61/94 (64%), Gaps = 5/94 (5%)
Query 37 EFVTYL-----RTYGRTTHFWGPVANWGFVIAGLSDMRKSPECISERMTGVLCIYSLLFM 91
F+T+L R Y +THFWGP++N+G IA + D++K P IS RMTG L +YS +FM
Sbjct 18 RFITWLKSPDFRKYLCSTHFWGPLSNFGIPIAAILDLKKDPRLISGRMTGALILYSSVFM 77
Query 92 RFATMVQPRNSLLFACHACNEAVQLTQLYRKLSY 125
R+A MV PRN LL CHA N VQ Q R +++
Sbjct 78 RYAWMVSPRNYLLLGCHAFNTTVQTAQGIRFVNF 111
> YGL080w
Length=130
Score = 92.8 bits (229), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 42/78 (53%), Positives = 51/78 (65%), Gaps = 0/78 (0%)
Query 48 TTHFWGPVANWGFVIAGLSDMRKSPECISERMTGVLCIYSLLFMRFATMVQPRNSLLFAC 107
TTHFWGPV+N+G IA + D++K P IS MT L YS +FM++A V P+N LLF C
Sbjct 28 TTHFWGPVSNFGIPIAAIYDLKKDPTLISGPMTFALVTYSGVFMKYALSVSPKNYLLFGC 87
Query 108 HACNEAVQLTQLYRKLSY 125
H NE QL Q YR L Y
Sbjct 88 HLINETAQLAQGYRFLKY 105
> Hs20549365
Length=323
Score = 91.7 bits (226), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 43/80 (53%), Positives = 49/80 (61%), Gaps = 0/80 (0%)
Query 42 LRTYGRTTHFWGPVANWGFVIAGLSDMRKSPECISERMTGVLCIYSLLFMRFATMVQPRN 101
R Y +THFWGP +WG +A DM+ SPE IS RMT L +YS +FMRFA VQPRN
Sbjct 207 FREYVSSTHFWGPAFSWGLPLAAFKDMKASPEIISGRMTTALILYSAIFMRFAYRVQPRN 266
Query 102 SLLFACHACNEAVQLTQLYR 121
LL ACH N Q Q R
Sbjct 267 LLLMACHCTNVMAQSVQASR 286
> Hs7661602
Length=127
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 44/80 (55%), Gaps = 0/80 (0%)
Query 47 RTTHFWGPVANWGFVIAGLSDMRKSPECISERMTGVLCIYSLLFMRFATMVQPRNSLLFA 106
RT FW P+ WG V AGL+DM + E +S + VL ++ R++ ++ P+N LFA
Sbjct 39 RTVFFWAPIMKWGLVCAGLADMARPAEKLSTAQSAVLMATGFIWSRYSLVIIPKNWSLFA 98
Query 107 CHACNEAVQLTQLYRKLSYN 126
+ A +QL+R YN
Sbjct 99 VNFFVGAAGASQLFRIWRYN 118
> At4g14690_2
Length=122
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 44/80 (55%), Gaps = 0/80 (0%)
Query 47 RTTHFWGPVANWGFVIAGLSDMRKSPECISERMTGVLCIYSLLFMRFATMVQPRNSLLFA 106
+T HFW P WG IA ++D +K PE IS + +++ R +T++ P+N LF+
Sbjct 30 KTIHFWAPTFKWGISIANIADFQKPPENISYLQQIAVTCTGMIWCRCSTIITPKNWNLFS 89
Query 107 CHACNEAVQLTQLYRKLSYN 126
+ A + QL RK+ Y+
Sbjct 90 VNVAMAATGIYQLTRKIKYD 109
> At4g22310
Length=108
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 26/91 (28%), Positives = 48/91 (52%), Gaps = 1/91 (1%)
Query 47 RTTHFWGPVANWGFVIAGLSDMRKSPECISERMTGVLCIYSLLFMRFATMVQPRNSLLFA 106
+T HFW P WG IA ++D K PE +S + +++ R++ ++ P+N LF+
Sbjct 17 KTIHFWAPTFKWGISIANIADFAKPPEKLSYPQQIAVTCTGVIWSRYSMVINPKNWNLFS 76
Query 107 CHACNEAVQLTQLYRKLSYN-ATKANSTTST 136
+ + QL RK+ ++ AT+A ++
Sbjct 77 VNVAMAGTGIYQLARKIKHDYATEAEPAVAS 107
> 7299210
Length=154
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 42/80 (52%), Gaps = 0/80 (0%)
Query 47 RTTHFWGPVANWGFVIAGLSDMRKSPECISERMTGVLCIYSLLFMRFATMVQPRNSLLFA 106
+T FW PV WG V AGLSD+ + + IS L +++ R++ ++ P+N LFA
Sbjct 58 KTIFFWAPVFKWGLVAAGLSDLARPADTISVSGCAALAATGIIWSRYSLVIIPKNYSLFA 117
Query 107 CHACNEAVQLTQLYRKLSYN 126
+ Q+ QL R Y+
Sbjct 118 VNLFVGITQVVQLARAYHYH 137
> YHR162w
Length=129
Score = 55.1 bits (131), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 29/88 (32%), Positives = 46/88 (52%), Gaps = 0/88 (0%)
Query 47 RTTHFWGPVANWGFVIAGLSDMRKSPECISERMTGVLCIYSLLFMRFATMVQPRNSLLFA 106
+T HFW P WG V AG SDM++ E IS L +L++ R++ +++PRN LL +
Sbjct 21 KTVHFWAPTLKWGLVFAGFSDMKRPVEKISGAQNLSLLSTALIWTRWSFVIKPRNILLAS 80
Query 107 CHACNEAVQLTQLYRKLSYNATKANSTT 134
++ QL R +Y +S +
Sbjct 81 VNSFLCLTAGYQLGRIANYRIRNGDSIS 108
> 7299209
Length=151
Score = 54.7 bits (130), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 43/80 (53%), Gaps = 0/80 (0%)
Query 47 RTTHFWGPVANWGFVIAGLSDMRKSPECISERMTGVLCIYSLLFMRFATMVQPRNSLLFA 106
+T FW P+ W VIAGLSD+ + + IS L +L++ R++ ++ P+N LFA
Sbjct 52 KTIFFWAPIVKWSLVIAGLSDLTRPADKISPNGCLALGATNLIWTRYSLVIIPKNYSLFA 111
Query 107 CHACNEAVQLTQLYRKLSYN 126
+ QL QL R +Y
Sbjct 112 VNLFVSLTQLFQLGRYYNYQ 131
> YGR243w
Length=146
Score = 54.3 bits (129), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 24/76 (31%), Positives = 43/76 (56%), Gaps = 3/76 (3%)
Query 47 RTTHFWGPVANWGFVIAGLSDMRKSPECISERMTGVLCIYSLLFMRFATMVQPRNSLLFA 106
+T HFW P WG V AGL+D+++ E +S L +L++ R++ +++P+N LL +
Sbjct 21 KTVHFWAPTLKWGLVFAGLNDIKRPVEKVSGAQNLSLLATALIWTRWSFVIKPKNYLLAS 80
Query 107 CH---ACNEAVQLTQL 119
+ C LT++
Sbjct 81 VNFFLGCTAGYHLTRI 96
> SPAC24B11.09
Length=118
Score = 54.3 bits (129), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 26/90 (28%), Positives = 43/90 (47%), Gaps = 0/90 (0%)
Query 47 RTTHFWGPVANWGFVIAGLSDMRKSPECISERMTGVLCIYSLLFMRFATMVQPRNSLLFA 106
+T HFW P W V++G+ D +SPE +S R LC ++ R++ +V+P+N
Sbjct 17 KTVHFWAPAMKWTLVLSGIGDYARSPEYLSIRQYAALCATGAIWTRWSLIVRPKNYFNAT 76
Query 107 CHACNEAVQLTQLYRKLSYNATKANSTTST 136
+ V Q+ R L Y + T +
Sbjct 77 VNFFLAIVGAVQVSRILVYQRQQKRITAQS 106
> At4g05590
Length=150
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/113 (23%), Positives = 44/113 (38%), Gaps = 38/113 (33%)
Query 49 THFWGPVANWGFVIAGLSDMRKSPECIS-------------------------------- 76
HFW P WG IA ++D +K PE +S
Sbjct 37 VHFWAPTFKWGISIANIADFQKPPETLSYPQQIGIILTIGLSYLYSAHIAVMYRCVNFNV 96
Query 77 ---ERMTGVLCIYS---LLFMRFATMVQPRNSLLFACHACNEAVQLTQLYRKL 123
RM ++ + + L++ R++T++ P+N LF+ + QL RK+
Sbjct 97 YMMPRMAVIVSVITGTGLVWSRYSTVITPKNWNLFSVSLGMAVTGIYQLTRKI 149
> CE28447
Length=133
Score = 35.8 bits (81), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 20/79 (25%), Positives = 34/79 (43%), Gaps = 0/79 (0%)
Query 47 RTTHFWGPVANWGFVIAGLSDMRKSPECISERMTGVLCIYSLLFMRFATMVQPRNSLLFA 106
+T FW P W + AGL+D+ + + +S L ++ R+ ++ P N L +
Sbjct 38 KTVFFWAPTIKWTLIGAGLADLARPADKLSLYQNSALFATGAIWTRYCLVITPINYYLSS 97
Query 107 CHACNEAVQLTQLYRKLSY 125
+ L QL R Y
Sbjct 98 VNFFVMCTGLAQLCRIAHY 116
> At1g14870
Length=152
Score = 30.8 bits (68), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 27/88 (30%), Positives = 37/88 (42%), Gaps = 13/88 (14%)
Query 51 FWGPVANWGFVIAGLSDMRKSPEC--------ISERMTGVLCIYSLLF---MRFATMVQP 99
FW P +G V A + D R S C + +TG CIYS + MR ++
Sbjct 34 FWCPCITFGQV-AEIVD-RGSTSCGTAGALYALIAVVTGCACIYSCFYRGKMRAQYNIKG 91
Query 100 RNSLLFACHACNEAVQLTQLYRKLSYNA 127
+ H C E LTQ YR+L +
Sbjct 92 DDCTDCLKHFCCELCSLTQQYRELKHRG 119
> 7296423
Length=1291
Score = 29.6 bits (65), Expect = 1.8, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 21/44 (47%), Gaps = 0/44 (0%)
Query 4 DVSEEPFRSAAFDASLPPVQGSLSATMASRGWSEFVTYLRTYGR 47
+ S RS+ F LP QG+ ++T A R E LR GR
Sbjct 989 NTSSLAHRSSTFYVDLPGFQGTATSTDAGRDEDELQRQLRQQGR 1032
> Hs21630255
Length=613
Score = 29.3 bits (64), Expect = 2.2, Method: Composition-based stats.
Identities = 11/13 (84%), Positives = 11/13 (84%), Gaps = 0/13 (0%)
Query 40 TYLRTYGRTTHFW 52
TYLRT GRTT FW
Sbjct 8 TYLRTLGRTTMFW 20
> 7291309
Length=1377
Score = 28.5 bits (62), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 18/69 (26%), Positives = 37/69 (53%), Gaps = 1/69 (1%)
Query 64 GLSDMRKSPECISERMTGVLCIYSLLFMRFATMVQPRNSLLFACHACNEAVQLTQLYRKL 123
GL + R S + ++ M +L + ++ +VQP+ ++ +C A N+A + +
Sbjct 672 GLLERRGSNQSLTLNMGSILGGAAGTEIQMTDIVQPKVTVC-SCAAANQAPHQRKFFSTE 730
Query 124 SYNATKANS 132
+ N++KANS
Sbjct 731 NLNSSKANS 739
> At4g34030
Length=630
Score = 27.3 bits (59), Expect = 9.4, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 27/60 (45%), Gaps = 1/60 (1%)
Query 29 TMASRGWSEFVTYLRTYGRTTHFWGPVANWGFVIAGLSDMRKSPECISERMTGVLCIYSL 88
T AS G + R Y F P A G ++ G + +P+C++ T C+Y+L
Sbjct 479 TGASFGAGNYAMCGRAYSPDFMFIWPNARIG-IMGGAQVNQNAPKCLNRNQTESKCVYNL 537
> Hs20471942
Length=571
Score = 27.3 bits (59), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 13/44 (29%), Positives = 23/44 (52%), Gaps = 2/44 (4%)
Query 32 SRGWSEFVTYLRTYGRTTHFWGPVANWGFVIAGLSDMRKSPECI 75
SR W++ + ++ G TT W P + + + L+ KSP C+
Sbjct 506 SRSWNDGLNSPKSVGLTTASWDPSVDLNWFL--LNQPNKSPSCL 547
Lambda K H
0.323 0.131 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1461889840
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40