bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3260_orf1
Length=155
Score E
Sequences producing significant alignments: (Bits) Value
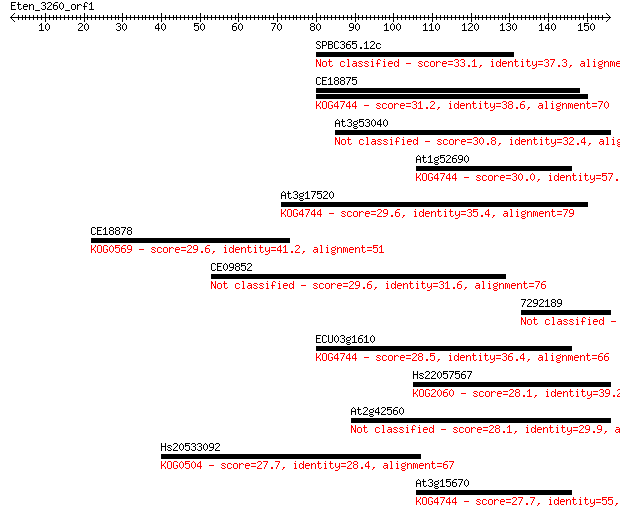
SPBC365.12c 33.1 0.20
CE18875 31.2 0.85
At3g53040 30.8 1.1
At1g52690 30.0 2.0
At3g17520 29.6 2.2
CE18878 29.6 2.3
CE09852 29.6 2.7
7292189 28.5 4.9
ECU03g1610 28.5 5.7
Hs22057567 28.1 6.6
At2g42560 28.1 6.6
Hs20533092 27.7 8.6
At3g15670 27.7 8.9
> SPBC365.12c
Length=684
Score = 33.1 bits (74), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 29/51 (56%), Gaps = 0/51 (0%)
Query 80 SARDSAVAAAQHSKEACCRAGADMKEKAGELGAAAKEKTGAAAEAAKDKAA 130
+A +S +A +H+K+ MK KA LG+ A EK G AA+ A + A+
Sbjct 389 TAAESVTSAFEHNKDTATSKAKQMKGKAASLGSTASEKVGEAADYAAETAS 439
> CE18875
Length=733
Score = 31.2 bits (69), Expect = 0.85, Method: Compositional matrix adjust.
Identities = 26/72 (36%), Positives = 39/72 (54%), Gaps = 8/72 (11%)
Query 80 SARDSAVAAAQHSKEACCRAGADMKEKAGELGAAAKEKTGAAAEAAKDKAAG----LQQR 135
SA+D A A +K+ A K+KAG+ K+K G A ++ KDKA+ + +
Sbjct 604 SAKDKASDAWDKTKDKAGEA----KDKAGDAWDNTKDKAGNAWDSTKDKASDAWDTTKDK 659
Query 136 SHEAYESAADAA 147
+ +A ESA DAA
Sbjct 660 ASDAKESAGDAA 671
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 37/70 (52%), Gaps = 4/70 (5%)
Query 80 SARDSAVAAAQHSKEACCRAGADMKEKAGELGAAAKEKTGAAAEAAKDKAAGLQQRSHEA 139
+A+D A A +K+ A K+K G+ K+K G A + KDKA + ++ +A
Sbjct 302 TAKDKASDAWDKTKDKAGEA----KDKMGDAWDTTKDKAGDAWDTTKDKAGDGKGKAGDA 357
Query 140 YESAADAAAD 149
+++ D A+D
Sbjct 358 WDTTKDKASD 367
> At3g53040
Length=479
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 23/79 (29%), Positives = 36/79 (45%), Gaps = 8/79 (10%)
Query 85 AVAAAQHSKEACCRAGAD--------MKEKAGELGAAAKEKTGAAAEAAKDKAAGLQQRS 136
AV H R GAD M++ GE+ + +KT A+ DKA + ++
Sbjct 68 AVIGKSHDTAESTREGADIASEKAAGMRDTTGEVRDSTAQKTKETADYTADKAREAKDKT 127
Query 137 HEAYESAADAAADKSEGAK 155
+ + AD AA+K+ AK
Sbjct 128 ADKTKETADYAAEKAREAK 146
> At1g52690
Length=169
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 23/51 (45%), Positives = 28/51 (54%), Gaps = 11/51 (21%)
Query 106 KAGELGAAAKEKTGAA-------AEAAKDK----AAGLQQRSHEAYESAAD 145
KAGE A+EKTG A +AAKDK A QQ++HE +SA D
Sbjct 10 KAGETRGKAQEKTGEAMGTMGDKTQAAKDKTQETAQSAQQKAHETAQSAKD 60
> At3g17520
Length=298
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 28/83 (33%), Positives = 45/83 (54%), Gaps = 8/83 (9%)
Query 71 SLQQTMSDPSARDSAVAAAQHSKEACCRAGADMKEKAGELGAAAKEKTGAAAEAAKDKA- 129
++Q T + + +D A A+Q +++A A K KA E AAKEK G+A E AK K
Sbjct 55 NIQPTHTTTTVQDDAWRASQKAEDAKEAA----KRKAEEAVGAAKEKAGSAYETAKSKVE 110
Query 130 ---AGLQQRSHEAYESAADAAAD 149
A ++ ++ ++Y+SA D
Sbjct 111 EGLASVKDKASQSYDSAGQVKDD 133
> CE18878
Length=479
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 21/59 (35%), Positives = 29/59 (49%), Gaps = 8/59 (13%)
Query 22 ISNFKVLLFQTF--------IQNSLNSFSSCCRAFSASVMLRFCCAAPLLLLLLLLLSL 72
IS F L QT + NSL SF+S A A++ + PLL+ LL+L+L
Sbjct 297 ISFFGTFLLQTIGFSPEGSAVANSLCSFASIVSALLAAIAMDKIGRRPLLITSLLVLAL 355
> CE09852
Length=96
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 24/76 (31%), Positives = 37/76 (48%), Gaps = 1/76 (1%)
Query 53 MLRFCCAAPLLLLLLLLLSLQQTMSDPSARDSAVAAAQHSKEACCRAGADMKEKAGELGA 112
M+R L++L LL + + Q +++ + D A A ++K A AD +G
Sbjct 1 MIRQTTTV-LVVLSLLAMFVAQEVAEEALTDPEAAEADNAKNNATDAPADATPGSGSDAP 59
Query 113 AAKEKTGAAAEAAKDK 128
AA E +GA AEA K
Sbjct 60 AAPEGSGAEAEATTAK 75
> 7292189
Length=516
Score = 28.5 bits (62), Expect = 4.9, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 133 QQRSHEAYESAADAAADKSEGAK 155
+QR+ + YESA DA AD +GA+
Sbjct 30 KQRNGKIYESAIDAVADVQDGAQ 52
> ECU03g1610
Length=166
Score = 28.5 bits (62), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 34/70 (48%), Gaps = 4/70 (5%)
Query 80 SARDSAVAAAQHSKEACCRAGADMKEKAGELGAAAKEKTGAAAEAAKDK----AAGLQQR 135
SA+D A +K+ K+K E +AK+KT AE+AKDK A + +
Sbjct 39 SAKDKTKETAGSAKDKTKETAESAKDKTKETAGSAKDKTKETAESAKDKTKETAGSAKDK 98
Query 136 SHEAYESAAD 145
+ E ESA D
Sbjct 99 TKETAESAKD 108
> Hs22057567
Length=1470
Score = 28.1 bits (61), Expect = 6.6, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 32/62 (51%), Gaps = 11/62 (17%)
Query 105 EKAGELGAAAKEKTGAAAEAAKDKAAGLQQRSHE------AYESAADAA-----ADKSEG 153
++ + G + TGA +E ++K A LQ+RS A S+ DAA D+S+G
Sbjct 185 QQTSQDGTLSDTATGAGSEVPREKKARLQERSRSQTPLSTAAASSQDAAPPSAPPDRSKG 244
Query 154 AK 155
A+
Sbjct 245 AE 246
> At2g42560
Length=635
Score = 28.1 bits (61), Expect = 6.6, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 29/67 (43%), Gaps = 0/67 (0%)
Query 89 AQHSKEACCRAGADMKEKAGELGAAAKEKTGAAAEAAKDKAAGLQQRSHEAYESAADAAA 148
AQ + E G + E A AK+ A AK+ AA QR+ E + AA
Sbjct 257 AQRASEYATEKGKEAGNMTAEQAARAKDYALQKAVEAKETAAEKAQRASEYMKETGSTAA 316
Query 149 DKSEGAK 155
+++ AK
Sbjct 317 EQAARAK 323
> Hs20533092
Length=161
Score = 27.7 bits (60), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 19/70 (27%), Positives = 37/70 (52%), Gaps = 6/70 (8%)
Query 40 SFSSCCRAFSASVMLRFCCAAPLLLLLLLLLSLQ---QTMSDPSARDSAVAAAQHSKEAC 96
+ ++ R +++L CC + ++ LLL ++ Q +S +AR+ AV++ H
Sbjct 74 NLNALDRYGRTALILAVCCGSASIVSLLLEQNIDVSSQDLSGQTAREYAVSSHHH---VI 130
Query 97 CRAGADMKEK 106
C+ +D KEK
Sbjct 131 CQLLSDYKEK 140
> At3g15670
Length=225
Score = 27.7 bits (60), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 22/51 (43%), Positives = 27/51 (52%), Gaps = 11/51 (21%)
Query 106 KAGELGAAAKEKTGAA-------AEAAKDK----AAGLQQRSHEAYESAAD 145
KAGE A+EKTG A AE +DK A QQ++HE +SA D
Sbjct 9 KAGETRGKAQEKTGQAMGTMRDKAEEGRDKTSQTAQTAQQKAHETAQSAKD 59
Lambda K H
0.318 0.124 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2033998736
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40