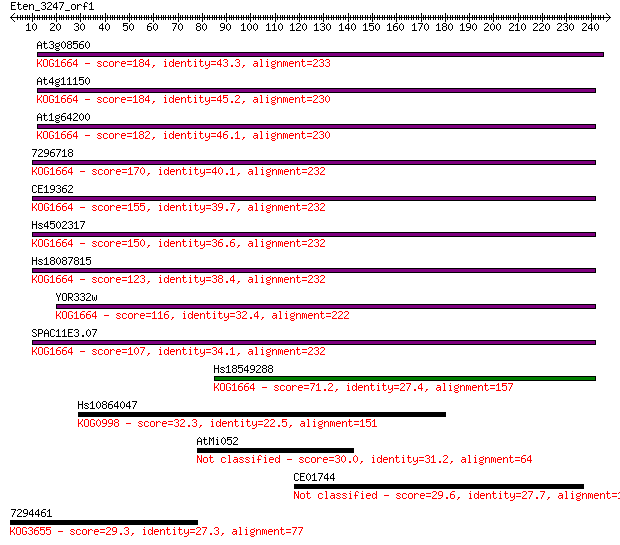
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3247_orf1
Length=247
Score E
Sequences producing significant alignments: (Bits) Value
At3g08560 184 2e-46
At4g11150 184 2e-46
At1g64200 182 7e-46
7296718 170 3e-42
CE19362 155 5e-38
Hs4502317 150 3e-36
Hs18087815 123 4e-28
YOR332w 116 4e-26
SPAC11E3.07 107 2e-23
Hs18549288 71.2 2e-12
Hs10864047 32.3 0.85
AtMi052 30.0 4.2
CE01744 29.6 6.3
7294461 29.3 7.5
> At3g08560
Length=235
Score = 184 bits (467), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 101/235 (42%), Positives = 150/235 (63%), Gaps = 7/235 (2%)
Query 12 MDDQEALAQIQQMVKFILNEAKDKAQEIEARALEDFNIEKLKLVQQMKDKIRQEFDKKAK 71
M+D + QIQQMV+FI EA++KA EI A E+FNIE+L+L++ K K+RQ++D+K K
Sbjct 1 MNDADVSKQIQQMVRFIRQEAEEKANEISISAEEEFNIERLQLLESAKRKLRQDYDRKLK 60
Query 72 KLEVQRSIDRSTAINKARLRRIAAQDQVVTEVYAQSQKQLATICSDTARYKELLTDLIVQ 131
++++++ ID ST +N +R++ + AQD VVT + + K L + +D YK+LL LI++
Sbjct 61 QVDIRKRIDYSTQLNASRIKYLQAQDDVVTAMKDSAAKDLLRVSNDKNNYKKLLKSLIIE 120
Query 132 GLLRLLEPEVVIRCREVDRSVVESVLPAAAAKYSKILNDEAGLKKTVKLSIDK--LGRYL 189
LLRL EP V++RCRE+D+ VVESV+ A +Y+ E + K++ID+
Sbjct 121 SLLRLKEPSVLLRCREMDKKVVESVIEDAKRQYA-----EKAKVGSPKITIDEKVFLPPP 175
Query 190 PPPPTADSTVPSCCGGVILVTADGRISCDNTLDARLKLVVTECAPAIRMHLFTTP 244
P P DS P C GGV+L + DG+I C+NTLDARL + + P IR L P
Sbjct 176 PNPKLPDSHDPHCSGGVVLASQDGKIVCENTLDARLDVAFRQKLPQIRTRLVGAP 230
> At4g11150
Length=230
Score = 184 bits (466), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 104/234 (44%), Positives = 145/234 (61%), Gaps = 13/234 (5%)
Query 12 MDDQEALAQIQQMVKFILNEAKDKAQEIEARALEDFNIEKLKLVQQMKDKIRQEFDKKAK 71
M+D + QIQQMV+FI EA++KA EI A E+FNIEKL+LV+ K KIRQ+++KK K
Sbjct 1 MNDGDVSRQIQQMVRFIRQEAEEKANEISVSAEEEFNIEKLQLVEAEKKKIRQDYEKKEK 60
Query 72 KLEVQRSIDRSTAINKARLRRIAAQDQVVTEVYAQSQKQLATICSDTARYKELLTDLIVQ 131
+ +V++ ID S +N +R++ + AQD +V + Q+ K L + D YK+LL DLIVQ
Sbjct 61 QADVRKKIDYSMQLNASRIKVLQAQDDIVNAMKDQAAKDLLNVSRDEYAYKQLLKDLIVQ 120
Query 132 GLLRLLEPEVVIRCREVDRSVVESVLPAAAAKY---SKILNDEAGLKKTVKLSIDKLGRY 188
LLRL EP V++RCRE D +VE+VL A +Y +K+ E + + +
Sbjct 121 CLLRLKEPSVLLRCREEDLGLVEAVLDDAKEEYAGKAKVHAPEVAVDTKI---------F 171
Query 189 LPPPP-TADSTVPSCCGGVILVTADGRISCDNTLDARLKLVVTECAPAIRMHLF 241
LPPPP + D C GGV+L + DG+I C+NTLDARL + P IR LF
Sbjct 172 LPPPPKSNDPHGLHCSGGVVLASRDGKIVCENTLDARLDVAFRMKLPVIRKSLF 225
> At1g64200
Length=237
Score = 182 bits (461), Expect = 7e-46, Method: Compositional matrix adjust.
Identities = 106/237 (44%), Positives = 150/237 (63%), Gaps = 13/237 (5%)
Query 12 MDDQEALAQIQQMVKFILNEAKDKAQEIEARALEDFNIEKLKLVQQMKDKIRQEFDKKAK 71
M+D +A QIQQMV+FI EA++KA EI + E+FNIEKL+LV+ K KIRQE++KK K
Sbjct 1 MNDADASIQIQQMVRFIRQEAEEKANEISISSEEEFNIEKLQLVEAEKKKIRQEYEKKEK 60
Query 72 KLEVQRSIDRSTAINKARLRRIAAQDQVVTEVYAQSQKQLATIC------SDTARYKELL 125
+++V++ ID S +N +R++ + AQD +V + ++ KQL + +YK LL
Sbjct 61 QVDVRKKIDYSMQLNASRIKVLQAQDDIVNAMKEEAAKQLLKVSQHGFFNHHHHQYKHLL 120
Query 126 TDLIVQGLLRLLEPEVVIRCREVDRSVVESVLPAAAAKYSKILNDEAGLKKTVKLSIDKL 185
DLIVQ LLRL EP V++RCRE D +VES+L A+ +Y K A ++ +DK
Sbjct 121 KDLIVQCLLRLKEPAVLLRCREEDLDIVESMLDDASEEYCKKAKVHAP-----EIIVDK- 174
Query 186 GRYLPPPPTADS-TVPSCCGGVILVTADGRISCDNTLDARLKLVVTECAPAIRMHLF 241
+LPP P+ D SC GGV+L + DG+I C+NTLDARL++ P IR LF
Sbjct 175 DIFLPPAPSDDDPHALSCAGGVVLASRDGKIVCENTLDARLEVAFRNKLPEIRKSLF 231
> 7296718
Length=226
Score = 170 bits (430), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 93/232 (40%), Positives = 146/232 (62%), Gaps = 16/232 (6%)
Query 10 MAMDDQEALAQIQQMVKFILNEAKDKAQEIEARALEDFNIEKLKLVQQMKDKIRQEFDKK 69
MA+ D + QI+ M+ FI EA +KA+EI+A+A E+FNIEK +LVQQ + KI + ++KK
Sbjct 1 MALSDADVQKQIKHMMAFIEQEANEKAEEIDAKAEEEFNIEKGRLVQQQRLKIMEYYEKK 60
Query 70 AKKLEVQRSIDRSTAINKARLRRIAAQDQVVTEVYAQSQKQLATICSDTARYKELLTDLI 129
K++E+Q+ I S +N+ARL+ + ++ V+ V ++K+L + + + Y+ +LT LI
Sbjct 61 EKQVELQKKIQSSNMLNQARLKVLKVREDHVSSVLDDARKRLGEVTKNQSEYETVLTKLI 120
Query 130 VQGLLRLLEPEVVIRCREVDRSVVESVLPAAAAKYSKILNDEAGLKKTVKLSIDKLGRYL 189
VQGL +++EP+V++RCREVD +V +VLPAA +Y +A + + V+L ID+
Sbjct 121 VQGLFQIMEPKVILRCREVDVPLVRNVLPAAVEQY------KAQINQNVELFIDE----- 169
Query 190 PPPPTADSTVPSCCGGVILVTADGRISCDNTLDARLKLVVTECAPAIRMHLF 241
D CGGV L+ +GRI NTL++RL L+ + P IR LF
Sbjct 170 -----KDFLSADTCGGVELLALNGRIKVPNTLESRLDLISQQLVPEIRNALF 216
> CE19362
Length=226
Score = 155 bits (393), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 92/232 (39%), Positives = 137/232 (59%), Gaps = 16/232 (6%)
Query 10 MAMDDQEALAQIQQMVKFILNEAKDKAQEIEARALEDFNIEKLKLVQQMKDKIRQEFDKK 69
M + D + Q++ M+ FI EA +KA+EI+A+A E+FNIEK +LVQQ + KI + F+KK
Sbjct 1 MGISDNDVQKQLRHMMAFIEQEANEKAEEIDAKAEEEFNIEKGRLVQQQRQKIMEFFEKK 60
Query 70 AKKLEVQRSIDRSTAINKARLRRIAAQDQVVTEVYAQSQKQLATICSDTARYKELLTDLI 129
K++E+QR I S ++N RLR + A++ + V +++ L+ I D ARY +L L+
Sbjct 61 EKQVELQRKIQASNSLNAGRLRCLKAREDHIGAVLDEARSNLSRISGDAARYPAILKGLV 120
Query 130 VQGLLRLLEPEVVIRCREVDRSVVESVLPAAAAKYSKILNDEAGLKKTVKLSIDKLGRYL 189
+QGLL+LLE EVV+RCRE D +VE +LP K E G T K+ +DK +L
Sbjct 121 MQGLLQLLEKEVVLRCREKDLRLVEQLLPECLDGLQK----EWG--STTKVVLDKQN-FL 173
Query 190 PPPPTADSTVPSCCGGVILVTADGRISCDNTLDARLKLVVTECAPAIRMHLF 241
P GGV L G+I +TL++RL+L+ + P +R LF
Sbjct 174 P---------SESAGGVELSARAGKIKVSSTLESRLELIANQIVPQVRTALF 216
> Hs4502317
Length=226
Score = 150 bits (378), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 85/232 (36%), Positives = 140/232 (60%), Gaps = 16/232 (6%)
Query 10 MAMDDQEALAQIQQMVKFILNEAKDKAQEIEARALEDFNIEKLKLVQQMKDKIRQEFDKK 69
MA+ D + QI+ M+ FI EA +KA+EI+A+A E+FNIEK +LVQ + KI + ++KK
Sbjct 1 MALSDADVQKQIKHMMAFIEQEANEKAEEIDAKAEEEFNIEKGRLVQTQRLKIMEYYEKK 60
Query 70 AKKLEVQRSIDRSTAINKARLRRIAAQDQVVTEVYAQSQKQLATICSDTARYKELLTDLI 129
K++E Q+ I S +N+ARL+ + A+D ++T++ +++++L+ + DT RY+ LL L+
Sbjct 61 EKQIEQQKKIQMSNLMNQARLKVLRARDDLITDLLNEAKQRLSKVVKDTTRYQVLLDGLV 120
Query 130 VQGLLRLLEPEVVIRCREVDRSVVESVLPAAAAKYSKILNDEAGLKKTVKLSIDKLGRYL 189
+QGL +LLEP +++RCR+ D +V++ + A Y + K V + ID+ YL
Sbjct 121 LQGLYQLLEPRMIVRCRKQDFPLVKAAVQKAIPMY------KIATKNDVDVQIDQES-YL 173
Query 190 PPPPTADSTVPSCCGGVILVTADGRISCDNTLDARLKLVVTECAPAIRMHLF 241
P GGV + D +I NTL++RL L+ + P +R LF
Sbjct 174 P---------EDIAGGVEIYNGDRKIKVSNTLESRLDLIAQQMMPEVRGALF 216
> Hs18087815
Length=226
Score = 123 bits (308), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 89/232 (38%), Positives = 142/232 (61%), Gaps = 16/232 (6%)
Query 10 MAMDDQEALAQIQQMVKFILNEAKDKAQEIEARALEDFNIEKLKLVQQMKDKIRQEFDKK 69
MA+ D + QI+ M+ FI EA +KA+EI+A+A E+FNIEK +LVQ + KI + ++KK
Sbjct 1 MALSDVDVKKQIKHMMAFIEQEANEKAEEIDAKAEEEFNIEKGRLVQTQRLKIMEYYEKK 60
Query 70 AKKLEVQRSIDRSTAINKARLRRIAAQDQVVTEVYAQSQKQLATICSDTARYKELLTDLI 129
K++E Q+ I ST N+ARL+ + A++ +++++ ++++ +L+ I D Y+ LL L+
Sbjct 61 EKQIEQQKKILMSTMRNQARLKVLRARNDLISDLLSEAKLRLSRIVEDPEVYQGLLDKLV 120
Query 130 VQGLLRLLEPEVVIRCREVDRSVVESVLPAAAAKYSKILNDEAGLKKTVKLSIDKLGRYL 189
+QGLLRLLEP +++RCR D +VE+ + A +Y I +K V++ IDK YL
Sbjct 121 LQGLLRLLEPVMIVRCRPQDLLLVEAAVQKAIPEYMTI------SQKHVEVQIDKEA-YL 173
Query 190 PPPPTADSTVPSCCGGVILVTADGRISCDNTLDARLKLVVTECAPAIRMHLF 241
+ GGV + + + RI NTL++RL L + P IRM LF
Sbjct 174 ---------AVNAAGGVEVYSGNQRIKVSNTLESRLDLSAKQKMPEIRMALF 216
> YOR332w
Length=233
Score = 116 bits (290), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 72/222 (32%), Positives = 129/222 (58%), Gaps = 14/222 (6%)
Query 20 QIQQMVKFILNEAKDKAQEIEARALEDFNIEKLKLVQQMKDKIRQEFDKKAKKLEVQRSI 79
++ +M FI EA++KA+EI+ +A +++ IEK +V+ + I F K KK + + I
Sbjct 16 ELNKMQAFIRKEAEEKAKEIQLKADQEYEIEKTNIVRNETNNIDGNFKSKLKKAMLSQQI 75
Query 80 DRSTAINKARLRRIAAQDQVVTEVYAQSQKQLATICSDTARYKELLTDLIVQGLLRLLEP 139
+ST NK RL+ ++A++Q + ++ +++++L+ I ++ YK +L LIV+ LL+LLEP
Sbjct 76 TKSTIANKMRLKVLSAREQSLDGIFEETKEKLSGIANNRDEYKPILQSLIVEALLKLLEP 135
Query 140 EVVIRCREVDRSVVESVLPAAAAKYSKILNDEAGLKKTVKLSIDKLGRYLPPPPTADSTV 199
+ +++ E D ++ES+ +Y + A L++ V +S D L + L
Sbjct 136 KAIVKALERDVDLIESMKDDIMREYGEKA-QRAPLEEIV-ISNDYLNKDL---------- 183
Query 200 PSCCGGVILVTADGRISCDNTLDARLKLVVTECAPAIRMHLF 241
GGV++ A +I +NTL+ RLKL+ E PAIR+ L+
Sbjct 184 --VSGGVVVSNASDKIEINNTLEERLKLLSEEALPAIRLELY 223
> SPAC11E3.07
Length=227
Score = 107 bits (267), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 79/232 (34%), Positives = 131/232 (56%), Gaps = 15/232 (6%)
Query 10 MAMDDQEALAQIQQMVKFILNEAKDKAQEIEARALEDFNIEKLKLVQQMKDKIRQEFDKK 69
M++ D++ A++ +MV FI EA +KA+EI + E+F +EK K+V++ D I Q +D K
Sbjct 1 MSLSDEQVQAEMHKMVSFIKQEALEKAKEIHTLSEEEFQVEKAKIVREQCDAIDQTYDMK 60
Query 70 AKKLEVQRSIDRSTAINKARLRRIAAQDQVVTEVYAQSQKQLATICSDTARYKELLTDLI 129
K+ + + I +S +NK+RL + ++ +V+ +++++ +K+L I Y + + DLI
Sbjct 61 LKRASMAQKIAKSNVLNKSRLEILNSKQKVIDDIFSRVEKKLDGIEQKKDAYTKFMADLI 120
Query 130 VQGLLRLLEPEVVIRCREVDRSVVESVLPAAAAKYSKILNDEAGLKKTVKLSIDKLGRYL 189
VQ + L EP ++ R+ D +V++ +P A +++L + G SID Y
Sbjct 121 VQAMELLGEPVGIVYSRQRDAEIVKAAIPKA----TEVLKSKNG-------SID----YE 165
Query 190 PPPPTADSTVPSCCGGVILVTADGRISCDNTLDARLKLVVTECAPAIRMHLF 241
T D S GGV+LV G+I DNTL ARL++V E P IR LF
Sbjct 166 LDAETDDFLNDSVLGGVVLVGLGGKIRVDNTLRARLEIVKEEALPEIRRLLF 217
> Hs18549288
Length=151
Score = 71.2 bits (173), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 43/157 (27%), Positives = 84/157 (53%), Gaps = 16/157 (10%)
Query 85 INKARLRRIAAQDQVVTEVYAQSQKQLATICSDTARYKELLTDLIVQGLLRLLEPEVVIR 144
+N+ARL+ + A+D ++T++ +++++L+ + DT RY+ LL L++QGL +LLE +++R
Sbjct 1 MNQARLKVLRARDDLITDLLNEAKQRLSKVVKDTTRYQVLLDGLVLQGLYQLLEHRMIVR 60
Query 145 CREVDRSVVESVLPAAAAKYSKILNDEAGLKKTVKLSIDKLGRYLPPPPTADSTVPSCCG 204
CR+ D +V++ + A Y + K V + ID+ YLP G
Sbjct 61 CRKQDLPLVKAAVQKAIPMY------KIATKNNVDVQIDQ-ESYLP---------EDIAG 104
Query 205 GVILVTADGRISCDNTLDARLKLVVTECAPAIRMHLF 241
GV + + +I T +++L L+ + ++ F
Sbjct 105 GVEIYNGNHKIKVSKTPESQLDLIAQQMMLEVQGAFF 141
> Hs10864047
Length=864
Score = 32.3 bits (72), Expect = 0.85, Method: Composition-based stats.
Identities = 34/154 (22%), Positives = 78/154 (50%), Gaps = 19/154 (12%)
Query 29 LNEAKDKAQEIEARALEDFNIEKLKLVQQMKDKIRQEFDKKAKKLEVQRSIDRSTAINKA 88
+ E D +QEI E +++E+ +++ ++ IRQ K ++ E+Q +DR T +
Sbjct 383 VKELDDISQEIAQLQREKYSLEQD--IREKEEAIRQ---KTSEVQELQNDLDRET----S 433
Query 89 RLRRIAAQDQVVTEVYAQSQKQLATICSDTARYKELLTDL--IVQGLLRLLEP-EVVIRC 145
L+ + AQ Q +Q +L + A+ +++L+D+ Q +++ + I+
Sbjct 434 SLQELEAQKQ-------DAQDRLDEMDQQKAKLRDMLSDVRQKCQDETQMISSLKTQIQS 486
Query 146 REVDRSVVESVLPAAAAKYSKILNDEAGLKKTVK 179
+E D E L A ++ +++ +E L+++++
Sbjct 487 QESDLKSQEDDLNRAKSELNRLQQEETQLEQSIQ 520
> AtMi052
Length=192
Score = 30.0 bits (66), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 31/64 (48%), Gaps = 2/64 (3%)
Query 78 SIDRSTAINKARLRRIAAQDQVVTEVYAQSQKQLATICSDTARYKELLTDLIVQGLLRLL 137
++ +T N RRI QD +VT+ Y KQ A C A E + + +V +LR++
Sbjct 129 NVKSATLTNATSSRRIRFQDDLVTKFYTLVGKQFAYSCISKAERVEFIRESLV--VLRMV 186
Query 138 EPEV 141
V
Sbjct 187 RGGV 190
> CE01744
Length=4385
Score = 29.6 bits (65), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 33/130 (25%), Positives = 55/130 (42%), Gaps = 21/130 (16%)
Query 118 TARYKELLTDLIVQGLLRLLEPEV---VIRCREVDRSVVESVLPAAAAKYSKILNDEAGL 174
T + +L T +VQ R+ V + ++D S ++ V P YS I ++ L
Sbjct 792 THHFVKLSTRDVVQAAFRISSESVDHLLTLVEKMDWSKIDLVEPV----YSNISSNVQAL 847
Query 175 KKTVKLSIDKLGRYLPPPPTADST--------VPSCCGGVILVTADGRISCDNTLDARLK 226
+ S LG+ LPP P ++T C VI + + I+ D +LD
Sbjct 848 ATSSSQSSGILGKLLPPAPEEETTSKEIREINTAKCILNVI-SSLNSTITSDASLD---- 902
Query 227 LVVTECAPAI 236
+++E AI
Sbjct 903 -MISESEQAI 911
> 7294461
Length=531
Score = 29.3 bits (64), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 42/77 (54%), Gaps = 12/77 (15%)
Query 1 SVYSHNTAKMAMDDQEALAQIQQMVKFILNEAKDKAQEIEARALEDFNIEKLKLVQQMKD 60
S YS + AM++Q+A + I +E+ A ++DF + ++ ++
Sbjct 137 SAYSFKEPRGAMEEQKAPVGTN-YTRVI------PTKELNASVMQDFWKK-----EEAEE 184
Query 61 KIRQEFDKKAKKLEVQR 77
K+RQE +K++K+LE+Q+
Sbjct 185 KLRQEAEKESKRLELQK 201
Lambda K H
0.318 0.131 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5009866206
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40