bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3190_orf3
Length=179
Score E
Sequences producing significant alignments: (Bits) Value
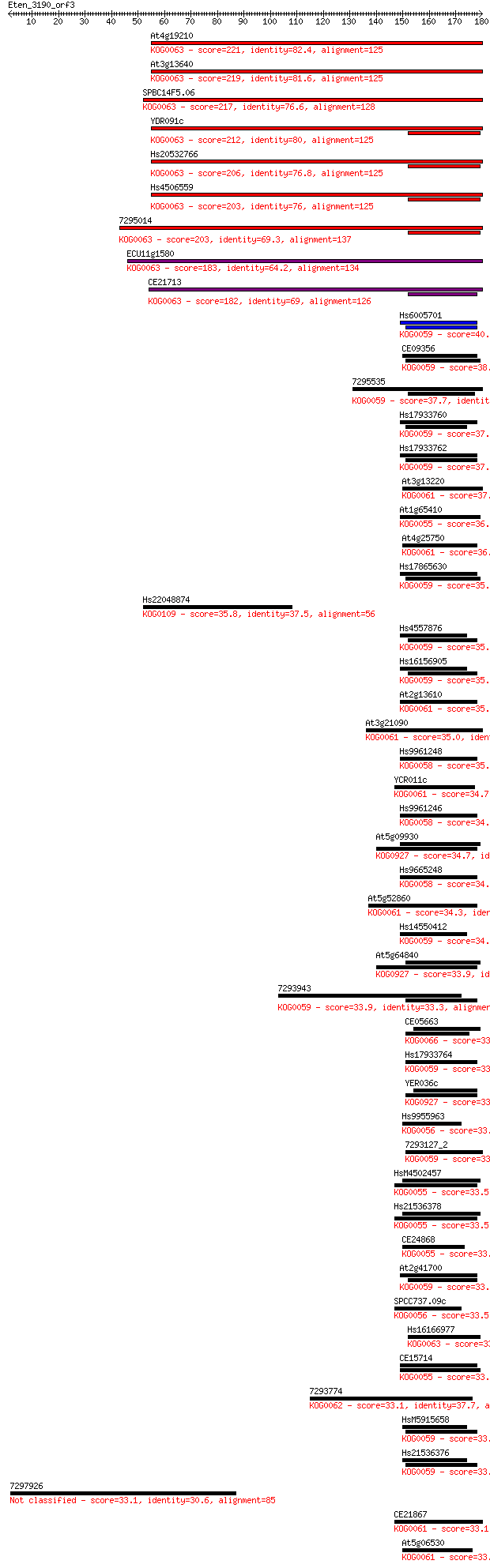
At4g19210 221 6e-58
At3g13640 219 2e-57
SPBC14F5.06 217 1e-56
YDR091c 212 3e-55
Hs20532766 206 3e-53
Hs4506559 203 1e-52
7295014 203 2e-52
ECU11g1580 183 2e-46
CE21713 182 4e-46
Hs6005701 40.8 0.002
CE09356 38.9 0.006
7295535 37.7 0.011
Hs17933760 37.7 0.012
Hs17933762 37.4 0.017
At3g13220 37.0 0.022
At1g65410 36.2 0.038
At4g25750 36.2 0.041
Hs17865630 35.8 0.042
Hs22048874 35.8 0.043
Hs4557876 35.8 0.050
Hs16156905 35.8 0.050
At2g13610 35.4 0.062
At3g21090 35.0 0.080
Hs9961248 35.0 0.086
YCR011c 34.7 0.093
Hs9961246 34.7 0.094
At5g09930 34.7 0.11
Hs9665248 34.7 0.12
At5g52860 34.3 0.12
Hs14550412 34.3 0.16
At5g64840 33.9 0.17
7293943 33.9 0.17
CE05663 33.9 0.19
Hs17933764 33.9 0.19
YER036c 33.5 0.20
Hs9955963 33.5 0.22
7293127_2 33.5 0.24
HsM4502457 33.5 0.25
Hs21536378 33.5 0.25
CE24868 33.5 0.25
At2g41700 33.5 0.25
SPCC737.09c 33.5 0.26
Hs16166977 33.5 0.27
CE15714 33.1 0.27
7293774 33.1 0.27
HsM5915658 33.1 0.30
Hs21536376 33.1 0.30
7297926 33.1 0.30
CE21867 33.1 0.33
At5g06530 33.1 0.33
> At4g19210
Length=600
Score = 221 bits (563), Expect = 6e-58, Method: Compositional matrix adjust.
Identities = 103/125 (82%), Positives = 109/125 (87%), Gaps = 0/125 (0%)
Query 55 RIAIVNGDKCKPKKCRQECKKSCPVVRTGKFCIEVESSSKVALISEPLCIGCGICVKKCP 114
RIAIV+ D+CKPKKCRQECKKSCPVV+TGK CIEV SK+A ISE LCIGCGICVKKCP
Sbjct 7 RIAIVSSDRCKPKKCRQECKKSCPVVKTGKLCIEVTVGSKLAFISEELCIGCGICVKKCP 66
Query 115 FSAIQIINLPRDLGAEVAHRYGPNQFKLHRLPVPRPGQVLGLVGTNGTGKSTALKILSAK 174
F AIQIINLPRDL + HRYG N FKLHRLPVPRPGQVLGLVGTNG GKSTALKIL+ K
Sbjct 67 FEAIQIINLPRDLEKDTTHRYGANTFKLHRLPVPRPGQVLGLVGTNGIGKSTALKILAGK 126
Query 175 LKPNL 179
LKPNL
Sbjct 127 LKPNL 131
> At3g13640
Length=603
Score = 219 bits (558), Expect = 2e-57, Method: Compositional matrix adjust.
Identities = 102/125 (81%), Positives = 110/125 (88%), Gaps = 0/125 (0%)
Query 55 RIAIVNGDKCKPKKCRQECKKSCPVVRTGKFCIEVESSSKVALISEPLCIGCGICVKKCP 114
RIAIV+ D+CKPKKCRQECKKSCPVV+TGK CIEV S+SK A ISE LCIGCGICVKKCP
Sbjct 7 RIAIVSEDRCKPKKCRQECKKSCPVVKTGKLCIEVGSTSKSAFISEELCIGCGICVKKCP 66
Query 115 FSAIQIINLPRDLGAEVAHRYGPNQFKLHRLPVPRPGQVLGLVGTNGTGKSTALKILSAK 174
F AIQIINLP+DL + HRYG N FKLHRLP+PRPGQVLGLVGTNG GKSTALKIL+ K
Sbjct 67 FEAIQIINLPKDLAKDTTHRYGANGFKLHRLPIPRPGQVLGLVGTNGIGKSTALKILAGK 126
Query 175 LKPNL 179
LKPNL
Sbjct 127 LKPNL 131
> SPBC14F5.06
Length=593
Score = 217 bits (552), Expect = 1e-56, Method: Composition-based stats.
Identities = 98/128 (76%), Positives = 108/128 (84%), Gaps = 0/128 (0%)
Query 52 SRLRIAIVNGDKCKPKKCRQECKKSCPVVRTGKFCIEVESSSKVALISEPLCIGCGICVK 111
S RIAIV+ DKC+PKKCRQEC++SCPVVRTGK CIEV + ++A ISE LCIGCGICVK
Sbjct 4 SLTRIAIVSEDKCRPKKCRQECRRSCPVVRTGKLCIEVNPTDRIAFISETLCIGCGICVK 63
Query 112 KCPFSAIQIINLPRDLGAEVAHRYGPNQFKLHRLPVPRPGQVLGLVGTNGTGKSTALKIL 171
KCPF AI IINLP +L +EV HRY N FKLHRLP PRPGQVLGLVGTNG GKSTALKIL
Sbjct 64 KCPFGAINIINLPTNLESEVTHRYSANSFKLHRLPTPRPGQVLGLVGTNGIGKSTALKIL 123
Query 172 SAKLKPNL 179
S K+KPNL
Sbjct 124 SGKMKPNL 131
> YDR091c
Length=608
Score = 212 bits (540), Expect = 3e-55, Method: Compositional matrix adjust.
Identities = 100/125 (80%), Positives = 107/125 (85%), Gaps = 0/125 (0%)
Query 55 RIAIVNGDKCKPKKCRQECKKSCPVVRTGKFCIEVESSSKVALISEPLCIGCGICVKKCP 114
RIAIV+ DKCKPKKCRQECK+SCPVV+TGK CIEV +SK+A ISE LCIGCGICVKKCP
Sbjct 7 RIAIVSADKCKPKKCRQECKRSCPVVKTGKLCIEVTPTSKIAFISEILCIGCGICVKKCP 66
Query 115 FSAIQIINLPRDLGAEVAHRYGPNQFKLHRLPVPRPGQVLGLVGTNGTGKSTALKILSAK 174
F AIQIINLP +L A V HRY N FKLHRLP PRPGQVLGLVGTNG GKSTALKIL+ K
Sbjct 67 FDAIQIINLPTNLEAHVTHRYSANSFKLHRLPTPRPGQVLGLVGTNGIGKSTALKILAGK 126
Query 175 LKPNL 179
KPNL
Sbjct 127 QKPNL 131
Score = 35.0 bits (79), Expect = 0.085, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 22/27 (81%), Gaps = 0/27 (0%)
Query 152 QVLGLVGTNGTGKSTALKILSAKLKPN 178
++L ++G NGTGK+T +K+L+ LKP+
Sbjct 379 EILVMMGENGTGKTTLIKLLAGALKPD 405
> Hs20532766
Length=599
Score = 206 bits (523), Expect = 3e-53, Method: Compositional matrix adjust.
Identities = 96/125 (76%), Positives = 104/125 (83%), Gaps = 0/125 (0%)
Query 55 RIAIVNGDKCKPKKCRQECKKSCPVVRTGKFCIEVESSSKVALISEPLCIGCGICVKKCP 114
RIAIVN DKCKPKKCRQECKKSCPVVR GK CIEV SK+A ISE LCIGCGIC+KKCP
Sbjct 7 RIAIVNHDKCKPKKCRQECKKSCPVVRMGKLCIEVTPQSKIAWISETLCIGCGICIKKCP 66
Query 115 FSAIQIINLPRDLGAEVAHRYGPNQFKLHRLPVPRPGQVLGLVGTNGTGKSTALKILSAK 174
F A+ I+NLP +L E HRY N FKLHRLP+PRPG+VLGLVGTNG GKSTALKIL+ K
Sbjct 67 FGALSIVNLPSNLEKETTHRYCANAFKLHRLPIPRPGEVLGLVGTNGIGKSTALKILAGK 126
Query 175 LKPNL 179
KPNL
Sbjct 127 QKPNL 131
Score = 34.3 bits (77), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 11/27 (40%), Positives = 23/27 (85%), Gaps = 0/27 (0%)
Query 152 QVLGLVGTNGTGKSTALKILSAKLKPN 178
+++ ++G NGTGK+T +++L+ +LKP+
Sbjct 373 EIMVMLGENGTGKTTFIRMLAGRLKPD 399
> Hs4506559
Length=599
Score = 203 bits (517), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 95/125 (76%), Positives = 103/125 (82%), Gaps = 0/125 (0%)
Query 55 RIAIVNGDKCKPKKCRQECKKSCPVVRTGKFCIEVESSSKVALISEPLCIGCGICVKKCP 114
RIAIVN DKCKPKKCRQECKKSCPVVR GK CIEV SK+A ISE LCIGCGIC+KKCP
Sbjct 7 RIAIVNHDKCKPKKCRQECKKSCPVVRMGKLCIEVTPQSKIAWISETLCIGCGICIKKCP 66
Query 115 FSAIQIINLPRDLGAEVAHRYGPNQFKLHRLPVPRPGQVLGLVGTNGTGKSTALKILSAK 174
F A+ I+NLP +L E HRY N FKLHRLP+PRPG+VLGLVGTNG GKS ALKIL+ K
Sbjct 67 FGALSIVNLPSNLEKETTHRYCANAFKLHRLPIPRPGEVLGLVGTNGIGKSAALKILAGK 126
Query 175 LKPNL 179
KPNL
Sbjct 127 QKPNL 131
Score = 34.3 bits (77), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 11/27 (40%), Positives = 23/27 (85%), Gaps = 0/27 (0%)
Query 152 QVLGLVGTNGTGKSTALKILSAKLKPN 178
+++ ++G NGTGK+T +++L+ +LKP+
Sbjct 373 EIMVMLGENGTGKTTFIRMLAGRLKPD 399
> 7295014
Length=611
Score = 203 bits (516), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 95/137 (69%), Positives = 109/137 (79%), Gaps = 0/137 (0%)
Query 43 KQQEGHEDPSRLRIAIVNGDKCKPKKCRQECKKSCPVVRTGKFCIEVESSSKVALISEPL 102
+++E E + RIAIV+ DKCKPK+CRQECKK+CPVVR GK CIEV +SK+A +SE L
Sbjct 3 RRKENEEVDKQTRIAIVSDDKCKPKRCRQECKKTCPVVRMGKLCIEVTPTSKIASLSEEL 62
Query 103 CIGCGICVKKCPFSAIQIINLPRDLGAEVAHRYGPNQFKLHRLPVPRPGQVLGLVGTNGT 162
CIGCGICVKKCPF AI IINLP +L HRY N FKLHRLP+PRPG+VLGLVG NG
Sbjct 63 CIGCGICVKKCPFEAITIINLPSNLEKHTTHRYSKNSFKLHRLPIPRPGEVLGLVGQNGI 122
Query 163 GKSTALKILSAKLKPNL 179
GKSTALKIL+ K KPNL
Sbjct 123 GKSTALKILAGKQKPNL 139
Score = 33.9 bits (76), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 22/27 (81%), Gaps = 0/27 (0%)
Query 152 QVLGLVGTNGTGKSTALKILSAKLKPN 178
++L L+G NGTGK+T +++L+ L+P+
Sbjct 381 EILVLLGENGTGKTTFIRMLAGNLQPD 407
> ECU11g1580
Length=624
Score = 183 bits (464), Expect = 2e-46, Method: Composition-based stats.
Identities = 86/134 (64%), Positives = 102/134 (76%), Gaps = 3/134 (2%)
Query 46 EGHEDPSRLRIAIVNGDKCKPKKCRQECKKSCPVVRTGKFCIEVESSSKVALISEPLCIG 105
+G E + R+AIVN + CKP +C EC+K+CPV +TGK CI +E K +ISE LCIG
Sbjct 3 KGEERTNLTRLAIVNFELCKPSQCASECRKACPVNKTGKECIRIE---KKCMISEVLCIG 59
Query 106 CGICVKKCPFSAIQIINLPRDLGAEVAHRYGPNQFKLHRLPVPRPGQVLGLVGTNGTGKS 165
C C KKCPF AI IINLP +L E+ HRYG N FKLHRLP+PRPG+VLGLVGTNG GKS
Sbjct 60 CSACQKKCPFKAISIINLPTNLTKEITHRYGTNGFKLHRLPIPRPGKVLGLVGTNGIGKS 119
Query 166 TALKILSAKLKPNL 179
TALKILS ++KPNL
Sbjct 120 TALKILSGEIKPNL 133
> CE21713
Length=610
Score = 182 bits (461), Expect = 4e-46, Method: Compositional matrix adjust.
Identities = 87/126 (69%), Positives = 99/126 (78%), Gaps = 0/126 (0%)
Query 54 LRIAIVNGDKCKPKKCRQECKKSCPVVRTGKFCIEVESSSKVALISEPLCIGCGICVKKC 113
LRIAIV D+CKPK C CK++CPV R GK CI VE++S ++ ISE LCIGCGICVKKC
Sbjct 17 LRIAIVEKDRCKPKNCGLACKRACPVNRQGKQCIVVEATSTISQISEILCIGCGICVKKC 76
Query 114 PFSAIQIINLPRDLGAEVAHRYGPNQFKLHRLPVPRPGQVLGLVGTNGTGKSTALKILSA 173
P+ AI+IINLP +L E HRY N FKLHRLP PR G+VLGLVGTNG GKSTALKIL+
Sbjct 77 PYDAIKIINLPANLANETTHRYSQNSFKLHRLPTPRCGEVLGLVGTNGIGKSTALKILAG 136
Query 174 KLKPNL 179
K KPNL
Sbjct 137 KQKPNL 142
Score = 32.7 bits (73), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 11/26 (42%), Positives = 21/26 (80%), Gaps = 0/26 (0%)
Query 152 QVLGLVGTNGTGKSTALKILSAKLKP 177
+++ ++G NGTGK+T +K+++ LKP
Sbjct 382 EIIVMLGENGTGKTTMIKMMAGSLKP 407
> Hs6005701
Length=1581
Score = 40.8 bits (94), Expect = 0.002, Method: Composition-based stats.
Identities = 16/29 (55%), Positives = 23/29 (79%), Gaps = 0/29 (0%)
Query 149 RPGQVLGLVGTNGTGKSTALKILSAKLKP 177
R G+VLGL+G NG GKST++K+++ KP
Sbjct 1274 RKGEVLGLLGHNGAGKSTSIKVITGDTKP 1302
Score = 28.9 bits (63), Expect = 6.1, Method: Composition-based stats.
Identities = 13/27 (48%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 151 GQVLGLVGTNGTGKSTALKILSAKLKP 177
GQ+ ++G +G GKST L ILS P
Sbjct 509 GQITAILGHSGAGKSTLLNILSGLSVP 535
> CE09356
Length=1447
Score = 38.9 bits (89), Expect = 0.006, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 150 PGQVLGLVGTNGTGKSTALKILSAKLKP 177
PG+ GL+G NG GK+T +L+AK++P
Sbjct 1185 PGECFGLLGLNGAGKTTTFAMLTAKIRP 1212
Score = 32.3 bits (72), Expect = 0.52, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 151 GQVLGLVGTNGTGKSTALKILSAKLKPN 178
GQ+ GL+G NG GK+T + IL P+
Sbjct 356 GQITGLLGHNGAGKTTTMSILCGLYAPS 383
> 7295535
Length=1500
Score = 37.7 bits (86), Expect = 0.011, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 30/49 (61%), Gaps = 1/49 (2%)
Query 131 VAHRYGPNQFKLHRLPVPRPGQVLGLVGTNGTGKSTALKILSAKLKPNL 179
+A+R G + V R G+ GL+G NG GKST K+L+ +L+P++
Sbjct 1160 LAYRRGHYAVRNVNFSVQR-GECFGLLGKNGAGKSTIFKLLTGQLQPDV 1207
Score = 29.3 bits (64), Expect = 4.1, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 19/25 (76%), Gaps = 0/25 (0%)
Query 152 QVLGLVGTNGTGKSTALKILSAKLK 176
QV L+G NG GKST +K+L+ +++
Sbjct 386 QVSCLLGRNGAGKSTLIKLLTGQIR 410
> Hs17933760
Length=1543
Score = 37.7 bits (86), Expect = 0.012, Method: Composition-based stats.
Identities = 15/29 (51%), Positives = 23/29 (79%), Gaps = 0/29 (0%)
Query 149 RPGQVLGLVGTNGTGKSTALKILSAKLKP 177
+ G+VLGL+G NG GKST++K+++ KP
Sbjct 1230 KKGEVLGLLGHNGAGKSTSIKMITGCTKP 1258
Score = 30.8 bits (68), Expect = 1.5, Method: Composition-based stats.
Identities = 13/23 (56%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 151 GQVLGLVGTNGTGKSTALKILSA 173
GQ+ ++G NG GKST L ILS
Sbjct 420 GQITAILGHNGAGKSTLLNILSG 442
> Hs17933762
Length=1624
Score = 37.4 bits (85), Expect = 0.017, Method: Composition-based stats.
Identities = 14/29 (48%), Positives = 22/29 (75%), Gaps = 0/29 (0%)
Query 149 RPGQVLGLVGTNGTGKSTALKILSAKLKP 177
+ G+V+GL+G NG GKST +K+++ KP
Sbjct 1317 KKGEVIGLLGHNGAGKSTTIKMITGDTKP 1345
Score = 28.9 bits (63), Expect = 6.5, Method: Composition-based stats.
Identities = 13/27 (48%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 151 GQVLGLVGTNGTGKSTALKILSAKLKP 177
GQ+ L+G +G GK+T L ILS P
Sbjct 510 GQITALLGHSGAGKTTLLNILSGLSVP 536
> At3g13220
Length=647
Score = 37.0 bits (84), Expect = 0.022, Method: Composition-based stats.
Identities = 14/30 (46%), Positives = 23/30 (76%), Gaps = 0/30 (0%)
Query 150 PGQVLGLVGTNGTGKSTALKILSAKLKPNL 179
PG++L L+G +G+GK+T LKI+ +L N+
Sbjct 78 PGEILALMGPSGSGKTTLLKIMGGRLTDNV 107
> At1g65410
Length=294
Score = 36.2 bits (82), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 23/30 (76%), Gaps = 0/30 (0%)
Query 149 RPGQVLGLVGTNGTGKSTALKILSAKLKPN 178
R G+ +G++G +GTGKST LKI++ L P+
Sbjct 38 RHGEAVGVIGPSGTGKSTILKIMAGLLAPD 67
> At4g25750
Length=577
Score = 36.2 bits (82), Expect = 0.041, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 20/28 (71%), Gaps = 0/28 (0%)
Query 150 PGQVLGLVGTNGTGKSTALKILSAKLKP 177
P Q+L ++G +G GKST L IL+A+ P
Sbjct 40 PSQILAIIGPSGAGKSTLLDILAARTSP 67
> Hs17865630
Length=1642
Score = 35.8 bits (81), Expect = 0.042, Method: Composition-based stats.
Identities = 14/29 (48%), Positives = 21/29 (72%), Gaps = 0/29 (0%)
Query 149 RPGQVLGLVGTNGTGKSTALKILSAKLKP 177
+ G++LGL+G NG GKST + IL ++P
Sbjct 1324 KKGEILGLLGPNGAGKSTIINILVGDIEP 1352
Score = 30.0 bits (66), Expect = 2.3, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 151 GQVLGLVGTNGTGKSTALKILSAKLKPN 178
GQ+ L+G +GTGKST + IL P+
Sbjct 507 GQITALLGHSGTGKSTLMNILCGLCPPS 534
> Hs22048874
Length=712
Score = 35.8 bits (81), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 31/59 (52%), Gaps = 3/59 (5%)
Query 52 SRLRIAIVNGDKCKPKKCRQE--CKKSCPVVRTGKFCIEVES-SSKVALISEPLCIGCG 107
+RLR A GD+C +C++E K CPV RTG+ E + + A + P +G G
Sbjct 428 ARLRTAPGMGDQCGCYQCKKEGHWSKECPVDRTGRVADFTEQCNEQYAAVHTPYTMGYG 486
> Hs4557876
Length=2273
Score = 35.8 bits (81), Expect = 0.050, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 149 RPGQVLGLVGTNGTGKSTALKILSA 173
RPG+ GL+G NG GK+T K+L+
Sbjct 1963 RPGECFGLLGVNGAGKTTTFKMLTG 1987
Score = 29.6 bits (65), Expect = 3.2, Method: Composition-based stats.
Identities = 12/26 (46%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 152 QVLGLVGTNGTGKSTALKILSAKLKP 177
Q+ +G NG GK+T L IL+ L P
Sbjct 957 QITAFLGHNGAGKTTTLSILTGLLPP 982
> Hs16156905
Length=2273
Score = 35.8 bits (81), Expect = 0.050, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 149 RPGQVLGLVGTNGTGKSTALKILSA 173
RPG+ GL+G NG GK+T K+L+
Sbjct 1963 RPGECFGLLGVNGAGKTTTFKMLTG 1987
Score = 29.6 bits (65), Expect = 3.2, Method: Composition-based stats.
Identities = 12/26 (46%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 152 QVLGLVGTNGTGKSTALKILSAKLKP 177
Q+ +G NG GK+T L IL+ L P
Sbjct 957 QITAFLGHNGAGKTTTLSILTGLLPP 982
> At2g13610
Length=649
Score = 35.4 bits (80), Expect = 0.062, Method: Composition-based stats.
Identities = 14/29 (48%), Positives = 23/29 (79%), Gaps = 0/29 (0%)
Query 149 RPGQVLGLVGTNGTGKSTALKILSAKLKP 177
+P ++L +VG +G GKS+ L+IL+A+L P
Sbjct 71 KPWEILAIVGPSGAGKSSLLEILAARLIP 99
> At3g21090
Length=680
Score = 35.0 bits (79), Expect = 0.080, Method: Composition-based stats.
Identities = 17/45 (37%), Positives = 28/45 (62%), Gaps = 1/45 (2%)
Query 136 GPNQFKLHRLP-VPRPGQVLGLVGTNGTGKSTALKILSAKLKPNL 179
GP + L RL PG+++ ++G +G+GKST L L+ +L N+
Sbjct 40 GPTRRLLQRLNGYAEPGRIMAIMGPSGSGKSTLLDSLAGRLARNV 84
> Hs9961248
Length=653
Score = 35.0 bits (79), Expect = 0.086, Method: Composition-based stats.
Identities = 15/29 (51%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 149 RPGQVLGLVGTNGTGKSTALKILSAKLKP 177
RPG+V LVG NG+GKST +L +P
Sbjct 494 RPGEVTALVGPNGSGKSTVAALLQNLYQP 522
> YCR011c
Length=1049
Score = 34.7 bits (78), Expect = 0.093, Method: Composition-based stats.
Identities = 14/30 (46%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 147 VPRPGQVLGLVGTNGTGKSTALKILSAKLK 176
+ +PGQ+L ++G +G GK+T L IL+ K K
Sbjct 412 IVKPGQILAIMGGSGAGKTTLLDILAMKRK 441
> Hs9961246
Length=703
Score = 34.7 bits (78), Expect = 0.094, Method: Composition-based stats.
Identities = 15/29 (51%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 149 RPGQVLGLVGTNGTGKSTALKILSAKLKP 177
RPG+V LVG NG+GKST +L +P
Sbjct 494 RPGEVTALVGPNGSGKSTVAALLQNLYQP 522
> At5g09930
Length=674
Score = 34.7 bits (78), Expect = 0.11, Method: Composition-based stats.
Identities = 12/30 (40%), Positives = 23/30 (76%), Gaps = 0/30 (0%)
Query 149 RPGQVLGLVGTNGTGKSTALKILSAKLKPN 178
+ G+ +GL+G NG GK+T L+I++ + +P+
Sbjct 107 KKGEKVGLIGVNGAGKTTQLRIITGQEEPD 136
Score = 29.3 bits (64), Expect = 4.3, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 22/38 (57%), Gaps = 1/38 (2%)
Query 140 FKLHRLPVPRPGQVLGLVGTNGTGKSTALKILSAKLKP 177
F L + R G+ + ++G NG GKST LK++ KP
Sbjct 422 FNKANLAIER-GEKVAIIGPNGCGKSTLLKLIMGLEKP 458
> Hs9665248
Length=808
Score = 34.7 bits (78), Expect = 0.12, Method: Composition-based stats.
Identities = 15/29 (51%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 149 RPGQVLGLVGTNGTGKSTALKILSAKLKP 177
RPG+V LVG NG+GKST +L +P
Sbjct 589 RPGEVTALVGPNGSGKSTVAALLQNLYQP 617
> At5g52860
Length=589
Score = 34.3 bits (77), Expect = 0.12, Method: Composition-based stats.
Identities = 17/42 (40%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 137 PNQFKLHRLPV-PRPGQVLGLVGTNGTGKSTALKILSAKLKP 177
P F L + + P ++L +VG +G GKST L IL++K P
Sbjct 40 PPSFILRNITLTAHPTEILAVVGPSGAGKSTLLDILASKTSP 81
> Hs14550412
Length=2436
Score = 34.3 bits (77), Expect = 0.16, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 149 RPGQVLGLVGTNGTGKSTALKILSA 173
RPG+ GL+G NG GK++ K+L+
Sbjct 2079 RPGECFGLLGVNGAGKTSTFKMLTG 2103
> At5g64840
Length=692
Score = 33.9 bits (76), Expect = 0.17, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 151 GQVLGLVGTNGTGKSTALKILSAKLKPN 178
G+ +GLVG NG GK+T L+I++ + +P+
Sbjct 123 GEKVGLVGVNGAGKTTQLRIITGQEEPD 150
Score = 30.0 bits (66), Expect = 2.4, Method: Composition-based stats.
Identities = 16/38 (42%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Query 140 FKLHRLPVPRPGQVLGLVGTNGTGKSTALKILSAKLKP 177
FK L + R G+ + ++G NG GKST LK++ KP
Sbjct 440 FKKANLSIER-GEKIAILGPNGCGKSTLLKLIMGLEKP 476
> 7293943
Length=1463
Score = 33.9 bits (76), Expect = 0.17, Method: Composition-based stats.
Identities = 24/90 (26%), Positives = 40/90 (44%), Gaps = 27/90 (30%)
Query 103 CIGCGICVKKCPFSAIQIINLP---------RDLGAE-----------VAHRYGPNQFKL 142
C GC CP+S+ I P R + A+ V+ +YG +Q +
Sbjct 1013 CKGC------CPYSSKGSIEDPKVSREAEKIRAMDADQIGTRALVVNGVSKKYGCDQLAV 1066
Query 143 HRLPVP-RPGQVLGLVGTNGTGKSTALKIL 171
+ + +P +GL+G NG GK+T K++
Sbjct 1067 NNISFALKPSHCVGLLGPNGAGKTTTFKMI 1096
Score = 28.1 bits (61), Expect = 8.9, Method: Composition-based stats.
Identities = 11/27 (40%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 151 GQVLGLVGTNGTGKSTALKILSAKLKP 177
GQ+ L+G NG GK+T + +L+ + P
Sbjct 364 GQITVLMGHNGAGKTTLISMLAGFISP 390
> CE05663
Length=622
Score = 33.9 bits (76), Expect = 0.19, Method: Composition-based stats.
Identities = 14/25 (56%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 154 LGLVGTNGTGKSTALKILSAKLKPN 178
+ +VG NG GKST LK+L K+ PN
Sbjct 433 IAIVGPNGVGKSTLLKLLIGKIDPN 457
Score = 28.9 bits (63), Expect = 5.2, Method: Composition-based stats.
Identities = 13/24 (54%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 151 GQVLGLVGTNGTGKSTALKILSAK 174
G+ GLVG NG GK+T LK + A+
Sbjct 105 GRRYGLVGPNGMGKTTLLKHIGAR 128
> Hs17933764
Length=1617
Score = 33.9 bits (76), Expect = 0.19, Method: Composition-based stats.
Identities = 12/27 (44%), Positives = 22/27 (81%), Gaps = 0/27 (0%)
Query 151 GQVLGLVGTNGTGKSTALKILSAKLKP 177
G++LGL+G +G GKS++++++S KP
Sbjct 1313 GEILGLLGPSGAGKSSSIRMISGITKP 1339
> YER036c
Length=610
Score = 33.5 bits (75), Expect = 0.20, Method: Composition-based stats.
Identities = 14/24 (58%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 154 LGLVGTNGTGKSTALKILSAKLKP 177
+ LVG NG GKST LKI++ +L P
Sbjct 424 IALVGPNGVGKSTLLKIMTGELTP 447
Score = 30.4 bits (67), Expect = 2.1, Method: Composition-based stats.
Identities = 14/27 (51%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 151 GQVLGLVGTNGTGKSTALKILSAKLKP 177
G+ GL+G NG GKST LK L+ + P
Sbjct 107 GRRYGLLGENGCGKSTFLKALATREYP 133
> Hs9955963
Length=842
Score = 33.5 bits (75), Expect = 0.22, Method: Composition-based stats.
Identities = 14/22 (63%), Positives = 17/22 (77%), Gaps = 0/22 (0%)
Query 150 PGQVLGLVGTNGTGKSTALKIL 171
PGQ L LVG +G GKST L++L
Sbjct 615 PGQTLALVGPSGAGKSTILRLL 636
> 7293127_2
Length=1586
Score = 33.5 bits (75), Expect = 0.24, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 21/29 (72%), Gaps = 0/29 (0%)
Query 151 GQVLGLVGTNGTGKSTALKILSAKLKPNL 179
G+V+ L+G N +GKST +K+L + +P L
Sbjct 439 GEVVALLGHNASGKSTIIKMLYGRTRPKL 467
> HsM4502457
Length=1321
Score = 33.5 bits (75), Expect = 0.25, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 150 PGQVLGLVGTNGTGKSTALKILSAKLKPN 178
PGQ L VG++G GKST++++L P+
Sbjct 1105 PGQTLAFVGSSGCGKSTSIQLLERFYDPD 1133
Score = 32.7 bits (73), Expect = 0.35, Method: Composition-based stats.
Identities = 14/31 (45%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 147 VPRPGQVLGLVGTNGTGKSTALKILSAKLKP 177
V +PG++ LVG +G GKSTAL+++ P
Sbjct 444 VIKPGEMTALVGPSGAGKSTALQLIQRFYDP 474
> Hs21536378
Length=1321
Score = 33.5 bits (75), Expect = 0.25, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 150 PGQVLGLVGTNGTGKSTALKILSAKLKPN 178
PGQ L VG++G GKST++++L P+
Sbjct 1105 PGQTLAFVGSSGCGKSTSIQLLERFYDPD 1133
Score = 32.7 bits (73), Expect = 0.35, Method: Composition-based stats.
Identities = 14/31 (45%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 147 VPRPGQVLGLVGTNGTGKSTALKILSAKLKP 177
V +PG++ LVG +G GKSTAL+++ P
Sbjct 444 VIKPGEMTALVGPSGAGKSTALQLIQRFYDP 474
> CE24868
Length=1239
Score = 33.5 bits (75), Expect = 0.25, Method: Composition-based stats.
Identities = 13/23 (56%), Positives = 19/23 (82%), Gaps = 0/23 (0%)
Query 150 PGQVLGLVGTNGTGKSTALKILS 172
PGQ + LVGT+G GKST++ +L+
Sbjct 426 PGQTVALVGTSGCGKSTSIGLLT 448
> At2g41700
Length=1850
Score = 33.5 bits (75), Expect = 0.25, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 149 RPGQVLGLVGTNGTGKSTALKILSAKLKP 177
+ G+ G +GTNG GK+T L +LS + P
Sbjct 1444 QAGECFGFLGTNGAGKTTTLSMLSGEETP 1472
Score = 30.8 bits (68), Expect = 1.6, Method: Composition-based stats.
Identities = 13/26 (50%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 152 QVLGLVGTNGTGKSTALKILSAKLKP 177
Q+L L+G NG GKST + +L L P
Sbjct 513 QILSLLGHNGAGKSTTISMLVGLLPP 538
> SPCC737.09c
Length=505
Score = 33.5 bits (75), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 14/25 (56%), Positives = 20/25 (80%), Gaps = 0/25 (0%)
Query 147 VPRPGQVLGLVGTNGTGKSTALKIL 171
V +PG+V+ LVG +G GKST ++IL
Sbjct 281 VAQPGKVIALVGESGGGKSTIMRIL 305
> Hs16166977
Length=124
Score = 33.5 bits (75), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 11/27 (40%), Positives = 22/27 (81%), Gaps = 0/27 (0%)
Query 152 QVLGLVGTNGTGKSTALKILSAKLKPN 178
+++ ++G NGTGK+T +++L+ +LKP
Sbjct 29 EIMVMLGENGTGKTTFIRMLAGRLKPE 55
> CE15714
Length=1294
Score = 33.1 bits (74), Expect = 0.27, Method: Composition-based stats.
Identities = 14/29 (48%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 149 RPGQVLGLVGTNGTGKSTALKILSAKLKP 177
+PGQ L LVG +G GKST + +L P
Sbjct 1056 KPGQTLALVGPSGCGKSTVISLLERLYDP 1084
Score = 33.1 bits (74), Expect = 0.29, Method: Composition-based stats.
Identities = 13/30 (43%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 149 RPGQVLGLVGTNGTGKSTALKILSAKLKPN 178
+PGQ + LVG++G GKST +++L P+
Sbjct 409 QPGQTVALVGSSGCGKSTIIQLLQRFYNPD 438
> 7293774
Length=708
Score = 33.1 bits (74), Expect = 0.27, Method: Composition-based stats.
Identities = 23/65 (35%), Positives = 32/65 (49%), Gaps = 5/65 (7%)
Query 115 FSAIQIINLPRDLGAEVAHRYGPNQ----FKLHRLPVPRPGQVLGLVGTNGTGKSTALKI 170
F ++ +N P +EV RY P FK L ++ +VG NG GKST LKI
Sbjct 480 FPEVEPLNPPVLAISEVTFRYNPEDPLPIFKGVNLSATSDSRIC-IVGENGAGKSTLLKI 538
Query 171 LSAKL 175
+ +L
Sbjct 539 IVGQL 543
> HsM5915658
Length=2201
Score = 33.1 bits (74), Expect = 0.30, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 150 PGQVLGLVGTNGTGKSTALKILSA 173
PG+ GL+G NG GKS+ K+L+
Sbjct 1878 PGECFGLLGVNGAGKSSTFKMLTG 1901
Score = 29.6 bits (65), Expect = 3.4, Method: Composition-based stats.
Identities = 11/27 (40%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 151 GQVLGLVGTNGTGKSTALKILSAKLKP 177
GQ+ +G NG GK+T + IL+ P
Sbjct 866 GQITSFLGHNGAGKTTTMSILTGLFPP 892
> Hs21536376
Length=2261
Score = 33.1 bits (74), Expect = 0.30, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 150 PGQVLGLVGTNGTGKSTALKILSA 173
PG+ GL+G NG GKS+ K+L+
Sbjct 1938 PGECFGLLGVNGAGKSSTFKMLTG 1961
Score = 29.6 bits (65), Expect = 3.4, Method: Composition-based stats.
Identities = 11/27 (40%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 151 GQVLGLVGTNGTGKSTALKILSAKLKP 177
GQ+ +G NG GK+T + IL+ P
Sbjct 926 GQITSFLGHNGAGKTTTMSILTGLFPP 952
> 7297926
Length=1354
Score = 33.1 bits (74), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 26/85 (30%), Positives = 37/85 (43%), Gaps = 8/85 (9%)
Query 2 HLKTKNKQFASLLSQQQQQIPQPRCSSRAAAAAAAAMAPGKKQQEGHEDPSRLRIAIVNG 61
+ KT NK F + SQ+ Q+ +R A A G+K + + P + I
Sbjct 216 YRKTSNKHFLRM-SQRLQR-------NRGCTVGAQFYAEGQKMRSDADKPCDICFCIRGT 267
Query 62 DKCKPKKCRQECKKSCPVVRTGKFC 86
+C PKKC K PVV G+ C
Sbjct 268 RRCAPKKCSPALKNCIPVVPKGQCC 292
> CE21867
Length=129
Score = 33.1 bits (74), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 147 VPRPGQVLGLVGTNGTGKSTALKILSAKLKPNL 179
V RPG+V ++G +G GK+T L +L+ + NL
Sbjct 88 VARPGEVTAIIGPSGAGKTTLLNVLTKRNLSNL 120
> At5g06530
Length=567
Score = 33.1 bits (74), Expect = 0.33, Method: Composition-based stats.
Identities = 12/26 (46%), Positives = 21/26 (80%), Gaps = 0/26 (0%)
Query 150 PGQVLGLVGTNGTGKSTALKILSAKL 175
PG+VL L+G +G+GK+T L +L+ ++
Sbjct 65 PGEVLALMGPSGSGKTTLLSLLAGRI 90
Lambda K H
0.319 0.134 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2815454148
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40