bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3177_orf1
Length=110
Score E
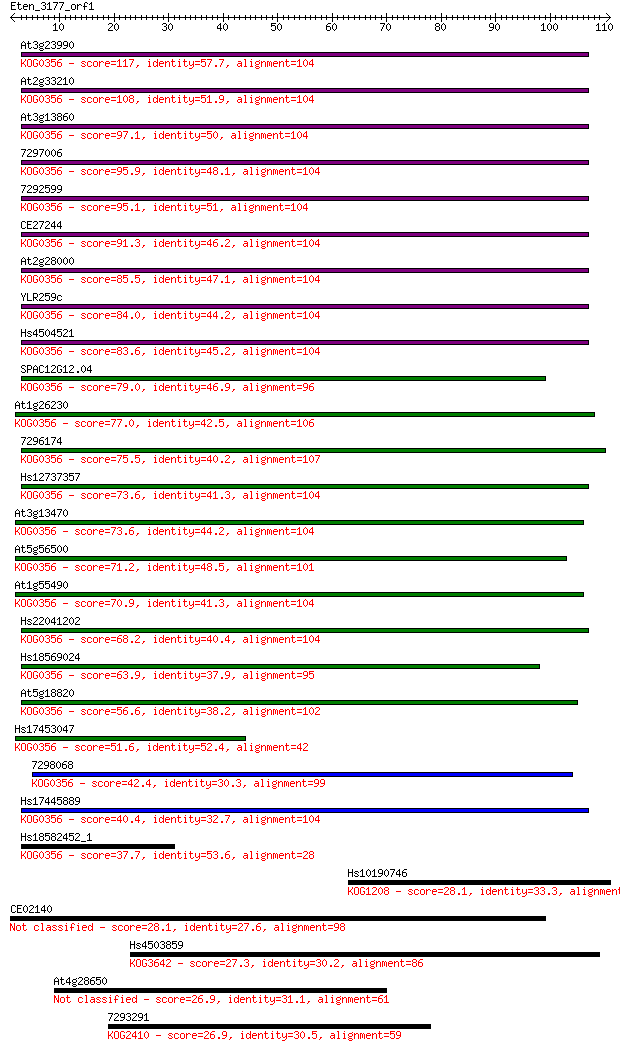
Sequences producing significant alignments: (Bits) Value
At3g23990 117 7e-27
At2g33210 108 2e-24
At3g13860 97.1 8e-21
7297006 95.9 1e-20
7292599 95.1 3e-20
CE27244 91.3 4e-19
At2g28000 85.5 2e-17
YLR259c 84.0 6e-17
Hs4504521 83.6 9e-17
SPAC12G12.04 79.0 2e-15
At1g26230 77.0 9e-15
7296174 75.5 2e-14
Hs12737357 73.6 8e-14
At3g13470 73.6 8e-14
At5g56500 71.2 5e-13
At1g55490 70.9 6e-13
Hs22041202 68.2 3e-12
Hs18569024 63.9 6e-11
At5g18820 56.6 1e-08
Hs17453047 51.6 4e-07
7298068 42.4 2e-04
Hs17445889 40.4 9e-04
Hs18582452_1 37.7 0.005
Hs10190746 28.1 4.2
CE02140 28.1 4.5
Hs4503859 27.3 6.3
At4g28650 26.9 8.5
7293291 26.9 9.1
> At3g23990
Length=577
Score = 117 bits (292), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 60/104 (57%), Positives = 82/104 (78%), Gaps = 2/104 (1%)
Query 3 EALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDLPSVGDEKLDIV 62
+ALATL++NKLR G++VCA+K+PGFG++RKA L DLA LTG +V+ D+L + EK+D+
Sbjct 287 DALATLILNKLRAGIKVCAIKAPGFGENRKANLQDLAALTGGEVITDEL-GMNLEKVDL- 344
Query 63 SMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIRTAIEQTKS 106
SMLG K V V+KD T++++GAG KK IEER QIR+AIE + S
Sbjct 345 SMLGTCKKVTVSKDDTVILDGAGDKKGIEERCEQIRSAIELSTS 388
> At2g33210
Length=524
Score = 108 bits (270), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 54/104 (51%), Positives = 79/104 (75%), Gaps = 2/104 (1%)
Query 3 EALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDLPSVGDEKLDIV 62
+ALATL++NKLR ++VCAVK+PGFG++RKA L+DLA LTGA+V+ ++L + + +D+
Sbjct 227 DALATLILNKLRANIKVCAVKAPGFGENRKANLHDLAALTGAQVITEEL-GMNLDNIDL- 284
Query 63 SMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIRTAIEQTKS 106
SM G K V V+KD T+V++GAG K+ I ER QIR+ +E + S
Sbjct 285 SMFGNCKKVTVSKDDTVVLDGAGDKQAIGERCEQIRSMVEASTS 328
> At3g13860
Length=554
Score = 97.1 bits (240), Expect = 8e-21, Method: Compositional matrix adjust.
Identities = 52/104 (50%), Positives = 75/104 (72%), Gaps = 2/104 (1%)
Query 3 EALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDLPSVGDEKLDIV 62
+ALA L++NK GL+VCA+K+PGFGD+RKA L DLA LTGA+V+ ++ + EK+
Sbjct 269 DALAMLILNKHHGGLKVCAIKAPGFGDNRKASLDDLAVLTGAEVISEE-RGLSLEKIR-P 326
Query 63 SMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIRTAIEQTKS 106
+LG AK V VT+D T+++ G G KK IEER ++R+A E++ S
Sbjct 327 ELLGTAKKVTVTRDDTIILHGGGDKKLIEERCEELRSANEKSTS 370
> 7297006
Length=573
Score = 95.9 bits (237), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 50/104 (48%), Positives = 73/104 (70%), Gaps = 1/104 (0%)
Query 3 EALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDLPSVGDEKLDIV 62
EAL+TLVVN+L++GL+VCAVK+PGFGD+RK L D+A TG V GD+ V E + +
Sbjct 278 EALSTLVVNRLKVGLQVCAVKAPGFGDNRKENLMDMAVATGGIVFGDEANMVRLEDIKM- 336
Query 63 SMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIRTAIEQTKS 106
S G+ V V+KD T++++G G K E+E+R +R AI+++ S
Sbjct 337 SDFGRVGEVVVSKDDTMLLKGKGQKAEVEKRVEGLREAIKESTS 380
> 7292599
Length=573
Score = 95.1 bits (235), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 53/104 (50%), Positives = 72/104 (69%), Gaps = 1/104 (0%)
Query 3 EALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDLPSVGDEKLDIV 62
EAL+TLVVN+L++GL+V AVK+PGFGD+RK+ L D+A +G V GDD V E + V
Sbjct 277 EALSTLVVNRLKIGLQVAAVKAPGFGDNRKSTLTDMAIASGGIVFGDDADLVKLEDVK-V 335
Query 63 SMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIRTAIEQTKS 106
S LG+ V +TKD TL+++G G K ++ RA QI+ IE T S
Sbjct 336 SDLGQVGEVVITKDDTLLLKGKGKKDDVLRRANQIKDQIEDTTS 379
> CE27244
Length=568
Score = 91.3 bits (225), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 48/104 (46%), Positives = 68/104 (65%), Gaps = 1/104 (0%)
Query 3 EALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDLPSVGDEKLDIV 62
EAL TLV+N+L++GL+V A+K+PGFGD+RK L D+ TGA + GDD + E +
Sbjct 272 EALTTLVLNRLKVGLQVVAIKAPGFGDNRKNTLKDMGIATGATIFGDDSNLIKIEDI-TA 330
Query 63 SMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIRTAIEQTKS 106
+ LG+ V +TKD TL++ G G + EIE+R I IEQ+ S
Sbjct 331 NDLGEVDEVTITKDDTLLLRGRGDQTEIEKRIEHITDEIEQSTS 374
> At2g28000
Length=586
Score = 85.5 bits (210), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 49/104 (47%), Positives = 69/104 (66%), Gaps = 2/104 (1%)
Query 3 EALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDLPSVGDEKLDIV 62
EALATLVVNKLR L V AVK+PGFG+ RKA L D+A LTGA+ + D+ + + +
Sbjct 301 EALATLVVNKLRGVLNVVAVKAPGFGERRKAMLQDIAILTGAEYLAMDMSLLVENA--TI 358
Query 63 SMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIRTAIEQTKS 106
LG A+ V ++KDST ++ A SK E++ R AQ++ + +T S
Sbjct 359 DQLGIARKVTISKDSTTLIADAASKDELQARIAQLKKELFETDS 402
> YLR259c
Length=572
Score = 84.0 bits (206), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 46/104 (44%), Positives = 71/104 (68%), Gaps = 2/104 (1%)
Query 3 EALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDLPSVGDEKLDIV 62
EALA ++NKLR ++VCAVK+PGFGD+RK + D+A LTG V ++L + E+ I
Sbjct 278 EALAACILNKLRGQVKVCAVKAPGFGDNRKNTIGDIAVLTGGTVFTEEL-DLKPEQCTIE 336
Query 63 SMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIRTAIEQTKS 106
+ LG S+ VTK+ T+++ G+G K+ I+ER QI+ +I+ T +
Sbjct 337 N-LGSCDSITVTKEDTVILNGSGPKEAIQERIEQIKGSIDITTT 379
> Hs4504521
Length=573
Score = 83.6 bits (205), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 47/104 (45%), Positives = 70/104 (67%), Gaps = 1/104 (0%)
Query 3 EALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDLPSVGDEKLDIV 62
EAL+TLV+N+L++GL+V AVK+PGFGD+RK QL D+A TG V G++ ++ E +
Sbjct 281 EALSTLVLNRLKVGLQVVAVKAPGFGDNRKNQLKDMAIATGGAVFGEEGLTLNLEDVQ-P 339
Query 63 SMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIRTAIEQTKS 106
LGK V VTKD ++++G G K +IE+R +I ++ T S
Sbjct 340 HDLGKVGEVIVTKDDAMLLKGKGDKAQIEKRIQEIIEQLDVTTS 383
> SPAC12G12.04
Length=582
Score = 79.0 bits (193), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 45/96 (46%), Positives = 62/96 (64%), Gaps = 2/96 (2%)
Query 3 EALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDLPSVGDEKLDIV 62
EALA ++NKLR L+V A+K+PGFGD+R+ L DLA LT + V D++ V EK
Sbjct 288 EALAACILNKLRGQLQVVAIKAPGFGDNRRNMLGDLAVLTDSAVFNDEI-DVSIEKAQ-P 345
Query 63 SMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIR 98
LG SV VTK+ T++M+GAG ++ +R QIR
Sbjct 346 HHLGSCGSVTVTKEDTIIMKGAGDHVKVNDRCEQIR 381
> At1g26230
Length=611
Score = 77.0 bits (188), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 45/110 (40%), Positives = 67/110 (60%), Gaps = 10/110 (9%)
Query 2 REALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDD----LPSVGDE 57
++ALA ++ NKL+ L+V A+K+P FG+ + L DLA TGA V+ D+ L G E
Sbjct 294 QDALAPVIRNKLKGNLKVAAIKAPAFGERKSHCLDDLAIFTGATVIRDEMGLSLEKAGKE 353
Query 58 KLDIVSMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIRTAIEQTKSG 107
+LG AK V VTKDSTL++ ++K ++ER +QI+ IE T+
Sbjct 354 ------VLGTAKRVLVTKDSTLIVTNGFTQKAVDERVSQIKNLIENTEEN 397
> 7296174
Length=648
Score = 75.5 bits (184), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 43/109 (39%), Positives = 69/109 (63%), Gaps = 5/109 (4%)
Query 3 EALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDL--PSVGDEKLD 60
EAL LV+NKLRLGL+VCAVKSP +G RK + D++ TGA + GDD+ + + KL+
Sbjct 275 EALNALVLNKLRLGLQVCAVKSPSYGHHRKELIGDISAATGATIFGDDINYSKMEEAKLE 334
Query 61 IVSMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIRTAIEQTKSGPK 109
+ +G+A ++KDST++++G +E R QI+ + + + P+
Sbjct 335 DLGQVGEAV---ISKDSTMLLQGKPKTGLLEMRIQQIQDELAEKQIKPE 380
> Hs12737357
Length=448
Score = 73.6 bits (179), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 43/104 (41%), Positives = 66/104 (63%), Gaps = 1/104 (0%)
Query 3 EALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDLPSVGDEKLDIV 62
EAL+TLV+N+L++GL+ A+K+PGFGD+RK QL D+A TG V G++ ++ E +
Sbjct 281 EALSTLVLNRLKVGLQDVAIKAPGFGDNRKNQLKDMAIATGGAVFGEEGLTLNLEDVQ-P 339
Query 63 SMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIRTAIEQTKS 106
LG V VTKD +++ G K +IE+R +I ++ T S
Sbjct 340 HDLGNVGEVIVTKDDATLLKEKGDKAQIEKRIQEIIEQLDVTTS 383
> At3g13470
Length=596
Score = 73.6 bits (179), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 46/108 (42%), Positives = 65/108 (60%), Gaps = 10/108 (9%)
Query 2 REALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKV----VGDDLPSVGDE 57
+EALATLVVNKLR L++ A+K+PGFG+ + L D+A LTGA V VG L G E
Sbjct 306 QEALATLVVNKLRGTLKIAALKAPGFGERKSQYLDDIAILTGATVIREEVGLSLDKAGKE 365
Query 58 KLDIVSMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIRTAIEQTK 105
+LG A V +TK+ T ++ +++ + +R QIR IEQ +
Sbjct 366 ------VLGNASKVVLTKEMTTIVGDGTTQEAVNKRVVQIRNLIEQAE 407
> At5g56500
Length=596
Score = 71.2 bits (173), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 49/106 (46%), Positives = 67/106 (63%), Gaps = 12/106 (11%)
Query 2 REALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKV----VGDDLPSVGDE 57
+E LATLVVNKLR ++V A+K+PGFG+ + L D+A LTGA V VG L VG E
Sbjct 306 QEPLATLVVNKLRGTIKVAALKAPGFGERKSQYLDDIAALTGATVIREEVGLQLEKVGPE 365
Query 58 KLDIVSMLGKAKSVEVTKDSTLVMEGAGSKKE-IEERAAQIRTAIE 102
+LG A V +TKD+T ++ G GS +E +++R QI+ IE
Sbjct 366 ------VLGNAGKVVLTKDTTTIV-GDGSTEEVVKKRVEQIKNLIE 404
> At1g55490
Length=600
Score = 70.9 bits (172), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 43/108 (39%), Positives = 66/108 (61%), Gaps = 10/108 (9%)
Query 2 REALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKV----VGDDLPSVGDE 57
+EALATLVVNKLR L++ A+++PGFG+ + L D+A LTGA V VG L G E
Sbjct 310 QEALATLVVNKLRGTLKIAALRAPGFGERKSQYLDDIAILTGATVIREEVGLSLDKAGKE 369
Query 58 KLDIVSMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIRTAIEQTK 105
+LG A V +TK+++ ++ ++ +++R QI+ IEQ +
Sbjct 370 ------VLGNASKVVLTKETSTIVGDGSTQDAVKKRVTQIKNLIEQAE 411
> Hs22041202
Length=485
Score = 68.2 bits (165), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 42/114 (36%), Positives = 66/114 (57%), Gaps = 11/114 (9%)
Query 3 EALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDLPSVGDEKLDIV 62
EAL+TLV+N+L++GL+V AVK PGFGD+R QL D+A TG V ++ ++ E +
Sbjct 230 EALSTLVLNRLKVGLQVVAVKDPGFGDNRNNQLKDMAIATGGAVFAEEGLTLNLEDVQ-P 288
Query 63 SMLGKAKSVEVTKDSTLVME----------GAGSKKEIEERAAQIRTAIEQTKS 106
LGK V VTKD ++++ G S E+ E+ ++ A+ T++
Sbjct 289 HDLGKVGEVIVTKDDAMLLKGKDGVAVLKVGGTSDAEVNEKQDRVTDALNATRA 342
> Hs18569024
Length=262
Score = 63.9 bits (154), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 36/95 (37%), Positives = 53/95 (55%), Gaps = 22/95 (23%)
Query 3 EALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDLPSVGDEKLDIV 62
EAL TL++N+L++GL+V AVK+PGFGD+RK QL D TG
Sbjct 123 EALTTLILNRLKVGLQVVAVKAPGFGDNRKNQLKDTVIATG------------------- 163
Query 63 SMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQI 97
G+ V V KD ++++G G+K +IE+ +I
Sbjct 164 ---GEVGEVTVIKDYAMLLKGKGNKSQIEKCVQEI 195
> At5g18820
Length=575
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 39/103 (37%), Positives = 58/103 (56%), Gaps = 4/103 (3%)
Query 3 EALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDLPSVGDEKLDIV 62
E L LVVNK + + V VK PG D +KA L D+A +TGA + DL G +
Sbjct 288 EVLEILVVNKKQGLINVAVVKCPGMLDGKKALLQDIALMTGADYLSGDL---GMSLMGAT 344
Query 63 S-MLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIRTAIEQT 104
S LG ++ V +T +ST ++ A +K EI+ R AQ++ + +T
Sbjct 345 SDQLGVSRRVVITANSTTIVADASTKPEIQARIAQMKKDLAET 387
> Hs17453047
Length=341
Score = 51.6 bits (122), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 22/42 (52%), Positives = 31/42 (73%), Gaps = 0/42 (0%)
Query 2 REALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTG 43
REA +T + N+L++GL++ AVK+P FGD+RK Q D A TG
Sbjct 214 REARSTFIFNRLKVGLQIIAVKAPDFGDNRKNQREDTAIATG 255
> 7298068
Length=558
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/101 (29%), Positives = 56/101 (55%), Gaps = 7/101 (6%)
Query 5 LATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGD--DLPSVGDEKLDIV 62
L LV+N L+ ++VCAVK+P FGD + ++ D+A TG ++ D L + +E L V
Sbjct 272 LKILVLNNLQGRVQVCAVKAPSFGDEQCEEMEDIAFATGGHLLEDASSLADLSEEDLGEV 331
Query 63 SMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIRTAIEQ 103
++V K++ L+ ++++++ R IR I++
Sbjct 332 -----MEAVVDAKETHLLQPINVNEEQVQCRIQDIRELIDE 367
> Hs17445889
Length=355
Score = 40.4 bits (93), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 34/104 (32%), Positives = 54/104 (51%), Gaps = 6/104 (5%)
Query 3 EALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDLPSVGDEKLDIV 62
EAL+TLV+N L++ + + GF K QL D+A T V G++ + E +
Sbjct 242 EALSTLVLNSLKIDFQS---RLQGF--VTKNQLKDMAVTTVGAVFGEEGLILHFEDVQ-P 295
Query 63 SMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIRTAIEQTKS 106
LGK V VTKD ++++G G +IE+ +I ++ T S
Sbjct 296 HDLGKVGEVIVTKDDAMLLKGKGGNAQIEKHIQEIIEQLDVTTS 339
> Hs18582452_1
Length=338
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 15/28 (53%), Positives = 23/28 (82%), Gaps = 0/28 (0%)
Query 3 EALATLVVNKLRLGLRVCAVKSPGFGDS 30
E L+TL++N+L++GL+V A K GFGD+
Sbjct 125 ETLSTLILNRLKVGLQVVAAKVSGFGDN 152
> Hs10190746
Length=336
Score = 28.1 bits (61), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query 63 SMLGKAKSVEVTKDSTLVMEGAGSKKEIEERAAQIRTAIEQ-TKSGPKP 110
S LG+A + E+ + V+ G + EE A Q+R + Q + GP+P
Sbjct 53 SGLGRATAAELLRLGARVIMGCRDRARAEEAAGQLRRELRQAAECGPEP 101
> CE02140
Length=233
Score = 28.1 bits (61), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 27/101 (26%), Positives = 42/101 (41%), Gaps = 21/101 (20%)
Query 1 RREALATLVVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDLPSVGDEKLD 60
R + A+L +NKL L C SP F LYD KV+ DD + ++
Sbjct 29 RDDPRASLALNKLHTALTTCEQASPSF-------LYDF-----TKVLLDD----SELSVN 72
Query 61 IVSMLGKAKSVEVTKDSTLVMEGA---GSKKEIEERAAQIR 98
+ + T D L++ G KE+ +RA ++R
Sbjct 73 LQESYLRMHDTSPTND--LIVSGYEQNADYKELTKRAIELR 111
> Hs4503859
Length=456
Score = 27.3 bits (59), Expect = 6.3, Method: Composition-based stats.
Identities = 26/86 (30%), Positives = 38/86 (44%), Gaps = 9/86 (10%)
Query 23 KSPGFGDSRKAQLYDLATLTGAKVVGDDLPSVGDEKLDIVSMLGKAKSVEVTKDSTLVME 82
KSPG D A + L+TLTG + G PS+ DE D ++ + + +
Sbjct 3 KSPGLSDCLWAWILLLSTLTG-RSYGQ--PSLQDELKDNTTVFTRILDRLLDGYDNRLRP 59
Query 83 GAGSKKEIEERAAQIRTAIEQTKSGP 108
G G ER +++T I T GP
Sbjct 60 GLG------ERVTEVKTDIFVTSFGP 79
> At4g28650
Length=1013
Score = 26.9 bits (58), Expect = 8.5, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 27/61 (44%), Gaps = 0/61 (0%)
Query 9 VVNKLRLGLRVCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDLPSVGDEKLDIVSMLGKA 68
V K+R + + P G+ R Q L L A + LP D++SMLG+A
Sbjct 920 VRRKIRDNISLEEALDPNVGNCRYVQEEMLLVLQIALLCTTKLPKDRPSMRDVISMLGEA 979
Query 69 K 69
K
Sbjct 980 K 980
> 7293291
Length=777
Score = 26.9 bits (58), Expect = 9.1, Method: Composition-based stats.
Identities = 18/63 (28%), Positives = 31/63 (49%), Gaps = 4/63 (6%)
Query 19 VCAVKSPGFGDSRKAQLYDLATLTGAKVVGDDLPSV----GDEKLDIVSMLGKAKSVEVT 74
V + SP FGD +A++ D L A+ G + ++ G L +++ G A SV +
Sbjct 548 VSRLNSPDFGDQNRAKINDSHVLPDAQAYGANFAAIEDLDGTSNLVVLAPNGDAVSVTSS 607
Query 75 KDS 77
+S
Sbjct 608 INS 610
Lambda K H
0.311 0.130 0.344
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1195973986
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40