bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3168_orf1
Length=175
Score E
Sequences producing significant alignments: (Bits) Value
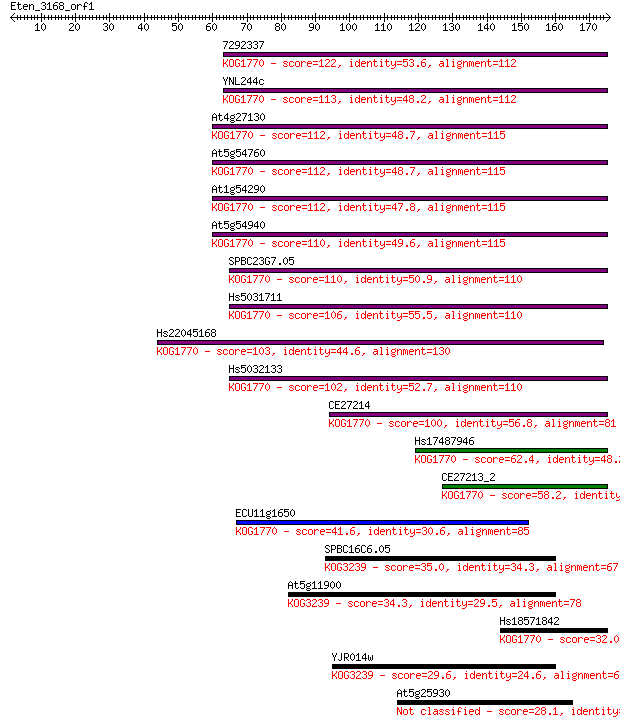
7292337 122 3e-28
YNL244c 113 2e-25
At4g27130 112 4e-25
At5g54760 112 4e-25
At1g54290 112 5e-25
At5g54940 110 2e-24
SPBC23G7.05 110 2e-24
Hs5031711 106 2e-23
Hs22045168 103 1e-22
Hs5032133 102 3e-22
CE27214 100 1e-21
Hs17487946 62.4 4e-10
CE27213_2 58.2 8e-09
ECU11g1650 41.6 7e-04
SPBC16C6.05 35.0 0.086
At5g11900 34.3 0.13
Hs18571842 32.0 0.57
YJR014w 29.6 3.2
At5g25930 28.1 9.6
> 7292337
Length=110
Score = 122 bits (307), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 60/112 (53%), Positives = 82/112 (73%), Gaps = 3/112 (2%)
Query 63 LDLQNLGVSDPFANDTTKAGGVGGNASTHLIHIRNQQRNGRKSVTTVQGLPKAFDLKKMV 122
+ +QNL DPFA D K G + L+HIR QQRNGRK++TTVQGL +DLKK+V
Sbjct 1 MSIQNLNTRDPFA-DAIK--GNDDDIQDGLVHIRIQQRNGRKTLTTVQGLSAEYDLKKIV 57
Query 123 RALKKEFSCNGTVIEDPEHGSIIQLQGDQRQAVKEFLEREGICGAEQLRIHG 174
R+ KKEF+CNGTVIE PE+G ++QLQGDQR+ + ++L + G+ +QL++HG
Sbjct 58 RSCKKEFACNGTVIEHPEYGEVLQLQGDQRENICQWLTKVGLAKPDQLKVHG 109
> YNL244c
Length=108
Score = 113 bits (282), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 54/112 (48%), Positives = 80/112 (71%), Gaps = 5/112 (4%)
Query 63 LDLQNLGVSDPFANDTTKAGGVGGNASTHLIHIRNQQRNGRKSVTTVQGLPKAFDLKKMV 122
+ ++NL DPFA+ G A+++ IHIR QQRNGRK++TTVQG+P+ +DLK+++
Sbjct 1 MSIENLKSFDPFAD-----TGDDETATSNYIHIRIQQRNGRKTLTTVQGVPEEYDLKRIL 55
Query 123 RALKKEFSCNGTVIEDPEHGSIIQLQGDQRQAVKEFLEREGICGAEQLRIHG 174
+ LKK+F+CNG +++DPE G IIQLQGDQR V EF+ + + ++IHG
Sbjct 56 KVLKKDFACNGNIVKDPEMGEIIQLQGDQRAKVCEFMISQLGLQKKNIKIHG 107
> At4g27130
Length=113
Score = 112 bits (280), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 56/115 (48%), Positives = 75/115 (65%), Gaps = 3/115 (2%)
Query 60 LMSLDLQNLGVSDPFANDTTKAGGVGGNASTHLIHIRNQQRNGRKSVTTVQGLPKAFDLK 119
+ LD Q DPFA+ + G G + +HIR QQRNGRKS+TTVQGL K +
Sbjct 1 MSELDSQVPTAFDPFADANAEDSGAG---TKEYVHIRVQQRNGRKSLTTVQGLKKEYSYT 57
Query 120 KMVRALKKEFSCNGTVIEDPEHGSIIQLQGDQRQAVKEFLEREGICGAEQLRIHG 174
K+++ LKKEF CNGTV++D E G +IQLQGDQR+ V FL + G+ + ++IHG
Sbjct 58 KILKDLKKEFCCNGTVVQDSELGQVIQLQGDQRKNVSTFLVQAGLVKKDNIKIHG 112
> At5g54760
Length=113
Score = 112 bits (280), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 56/115 (48%), Positives = 75/115 (65%), Gaps = 3/115 (2%)
Query 60 LMSLDLQNLGVSDPFANDTTKAGGVGGNASTHLIHIRNQQRNGRKSVTTVQGLPKAFDLK 119
+ LD Q DPFA+ + G G + +HIR QQRNGRKS+TTVQGL K +
Sbjct 1 MSELDSQVPTAFDPFADANVEDSGAG---TKEYVHIRVQQRNGRKSLTTVQGLKKEYSYT 57
Query 120 KMVRALKKEFSCNGTVIEDPEHGSIIQLQGDQRQAVKEFLEREGICGAEQLRIHG 174
K+++ LKKEF CNGTV++D E G +IQLQGDQR+ V FL + G+ + ++IHG
Sbjct 58 KILKDLKKEFCCNGTVVQDSELGQVIQLQGDQRKNVSTFLVQAGLVKKDNIKIHG 112
> At1g54290
Length=113
Score = 112 bits (279), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 55/115 (47%), Positives = 76/115 (66%), Gaps = 3/115 (2%)
Query 60 LMSLDLQNLGVSDPFANDTTKAGGVGGNASTHLIHIRNQQRNGRKSVTTVQGLPKAFDLK 119
+ L++Q DPFA+ + G G + +HIR QQRNGRKS+TTVQGL K +
Sbjct 1 MSDLEVQVPTAFDPFADANAEDSGAG---TKEYVHIRVQQRNGRKSLTTVQGLKKEYSYS 57
Query 120 KMVRALKKEFSCNGTVIEDPEHGSIIQLQGDQRQAVKEFLEREGICGAEQLRIHG 174
K+++ LKKEF CNGTV++D E G +IQLQGDQR+ V FL + G+ + ++IHG
Sbjct 58 KILKDLKKEFCCNGTVVQDSELGQVIQLQGDQRKNVSTFLVQAGLVKKDNIKIHG 112
> At5g54940
Length=112
Score = 110 bits (275), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 57/116 (49%), Positives = 78/116 (67%), Gaps = 6/116 (5%)
Query 60 LMSLDLQNLGVSDPFAN-DTTKAGGVGGNASTHLIHIRNQQRNGRKSVTTVQGLPKAFDL 118
++ LD+Q DPFA + A G N IHIR QQRNG+KS+TTVQGL K +
Sbjct 1 MVELDIQIPSAYDPFAEAKDSDAPGAKEN-----IHIRIQQRNGKKSLTTVQGLKKEYSY 55
Query 119 KKMVRALKKEFSCNGTVIEDPEHGSIIQLQGDQRQAVKEFLEREGICGAEQLRIHG 174
+++++ LKK+F CNG V++D E G IIQLQGDQR+ V +FL + GI +Q++IHG
Sbjct 56 ERILKDLKKDFCCNGNVVQDKELGKIIQLQGDQRKKVSQFLVQTGIAKKDQIKIHG 111
> SPBC23G7.05
Length=109
Score = 110 bits (275), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 56/112 (50%), Positives = 78/112 (69%), Gaps = 9/112 (8%)
Query 65 LQNLGVSDPFANDTTKAGGVGGNASTHL--IHIRNQQRNGRKSVTTVQGLPKAFDLKKMV 122
+QN DPFA+ G + + L IHIR QQRNGRK++TTVQGLP+ FD K+++
Sbjct 4 IQNFNTVDPFAD-------TGDDDAQPLNNIHIRIQQRNGRKTLTTVQGLPREFDQKRIL 56
Query 123 RALKKEFSCNGTVIEDPEHGSIIQLQGDQRQAVKEFLEREGICGAEQLRIHG 174
RALKK+F+CNGT+++D + G +IQLQGDQR V EFL ++ + ++IHG
Sbjct 57 RALKKDFACNGTIVKDDDLGEVIQLQGDQRIKVMEFLTQQLGLQKKNIKIHG 108
> Hs5031711
Length=113
Score = 106 bits (265), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 61/110 (55%), Positives = 79/110 (71%), Gaps = 1/110 (0%)
Query 65 LQNLGVSDPFANDTTKAGGVGGNASTHLIHIRNQQRNGRKSVTTVQGLPKAFDLKKMVRA 124
+QNL DPFA D TK + + IHIR QQRNGRK++TTVQG+ +D KK+V+A
Sbjct 4 IQNLQSFDPFA-DATKGDDLLPAGTEDYIHIRIQQRNGRKTLTTVQGIADDYDKKKLVKA 62
Query 125 LKKEFSCNGTVIEDPEHGSIIQLQGDQRQAVKEFLEREGICGAEQLRIHG 174
KK+F+CNGTVIE PE+G +IQLQGDQR+ + +FL GI EQL++HG
Sbjct 63 FKKKFACNGTVIEHPEYGEVIQLQGDQRKNICQFLLEVGIVKEEQLKVHG 112
> Hs22045168
Length=287
Score = 103 bits (258), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 58/130 (44%), Positives = 81/130 (62%), Gaps = 3/130 (2%)
Query 44 PKVGIHCVLHQREADTLMSLDLQNLGVSDPFANDTTKAGGVGGNASTHLIHIRNQQRNGR 103
P+V V + E ++ +QNL DPFA D +K + + IHIR QQRNGR
Sbjct 159 PQVPTQAVSTE-EKESYCKFAIQNLHSFDPFA-DASKGDDLLPAGTEDYIHIRIQQRNGR 216
Query 104 KSVTTVQGLPKAFDLKKMVRALKKEFSCNGTVIEDPEHGSIIQLQGDQRQAVKEFLEREG 163
K++TTVQG+ +D KK+V+A K F+CNGTVIE PE+G +IQ QGDQ + +FL G
Sbjct 217 KTLTTVQGIADDYDKKKLVKAFKN-FACNGTVIEHPEYGEVIQPQGDQCNNICQFLVELG 275
Query 164 ICGAEQLRIH 173
+ +QL++H
Sbjct 276 LAKDDQLKVH 285
> Hs5032133
Length=113
Score = 102 bits (255), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 58/110 (52%), Positives = 79/110 (71%), Gaps = 1/110 (0%)
Query 65 LQNLGVSDPFANDTTKAGGVGGNASTHLIHIRNQQRNGRKSVTTVQGLPKAFDLKKMVRA 124
+QNL DPFA D +K + + IHIR QQRNGRK++TTVQG+ +D KK+V+A
Sbjct 4 IQNLHSFDPFA-DASKGDDLLPAGTEDYIHIRIQQRNGRKTLTTVQGIADDYDKKKLVKA 62
Query 125 LKKEFSCNGTVIEDPEHGSIIQLQGDQRQAVKEFLEREGICGAEQLRIHG 174
KK+F+CNGTVIE PE+G +IQLQGDQR+ + +FL G+ +QL++HG
Sbjct 63 FKKKFACNGTVIEHPEYGEVIQLQGDQRKNICQFLVEIGLAKDDQLKVHG 112
> CE27214
Length=109
Score = 100 bits (250), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 46/81 (56%), Positives = 61/81 (75%), Gaps = 0/81 (0%)
Query 94 HIRNQQRNGRKSVTTVQGLPKAFDLKKMVRALKKEFSCNGTVIEDPEHGSIIQLQGDQRQ 153
HIR QQR GRK++TTVQG+ +DLK++V+ LKK+ SCNGT++E PE+G +IQL GDQR
Sbjct 28 HIRIQQRTGRKTITTVQGIGTEYDLKRIVQYLKKKHSCNGTIVEHPEYGEVIQLTGDQRD 87
Query 154 AVKEFLEREGICGAEQLRIHG 174
VK+FL + GI R+HG
Sbjct 88 KVKDFLIKVGIVNESNCRVHG 108
> Hs17487946
Length=113
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 27/56 (48%), Positives = 41/56 (73%), Gaps = 0/56 (0%)
Query 119 KKMVRALKKEFSCNGTVIEDPEHGSIIQLQGDQRQAVKEFLEREGICGAEQLRIHG 174
+++V+A K++F CNGTVIE PE+G +IQLQGDQR+ + +FL G+ + L + G
Sbjct 57 QQLVKAFKRKFPCNGTVIEHPEYGEVIQLQGDQRKNICQFLVEVGLAKDDLLMVQG 112
> CE27213_2
Length=49
Score = 58.2 bits (139), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 25/48 (52%), Positives = 34/48 (70%), Gaps = 0/48 (0%)
Query 127 KEFSCNGTVIEDPEHGSIIQLQGDQRQAVKEFLEREGICGAEQLRIHG 174
++ SCNGT++E PE+G +IQL GDQR VK+FL + GI R+HG
Sbjct 1 QKHSCNGTIVEHPEYGEVIQLTGDQRDKVKDFLIKVGIVNESNCRVHG 48
> ECU11g1650
Length=123
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 45/89 (50%), Gaps = 8/89 (8%)
Query 67 NLGVSDPFANDTTKAGGVGG----NASTHLIHIRNQQRNGRKSVTTVQGLPKAFDLKKMV 122
N D ++ +AG G +I ++ QQ+ G K VT ++ +P + ++
Sbjct 14 NAKEEDLSTSENPRAGQYGERDIIQEEKSVIRVKVQQKKGTKKVTIIENIPMEL-RESLL 72
Query 123 RALKKEFSCNGTVIEDPEHGSIIQLQGDQ 151
+LK+E C G +++D S IQLQGD+
Sbjct 73 SSLKREMGCGGILLDDK---SSIQLQGDK 98
> SPBC16C6.05
Length=190
Score = 35.0 bits (79), Expect = 0.086, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 37/69 (53%), Gaps = 3/69 (4%)
Query 93 IHIRNQQRNGRKSVTTVQGLPKAF--DLKKMVRALKKEFSCNGTVIEDPEHGSIIQLQGD 150
+ I+ +R RK VTTVQGL AF + KK + L +F+ +V + + I +QGD
Sbjct 99 VLIKTIERTKRKRVTTVQGLD-AFGIETKKAAKMLANKFATGASVTKTADKKDEIVVQGD 157
Query 151 QRQAVKEFL 159
+ +F+
Sbjct 158 LNYDIFDFI 166
> At5g11900
Length=228
Score = 34.3 bits (77), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 35/79 (44%), Gaps = 1/79 (1%)
Query 82 GGVGGNASTHLIHIRNQQRNGRKSVTTVQGLPK-AFDLKKMVRALKKEFSCNGTVIEDPE 140
GG + I RN RK +T V+GL L + L K+F+ +V++ P
Sbjct 126 GGKVKKKDRQEVIIEKVVRNKRKCITIVKGLELFGIKLSDASKKLGKKFATGASVVKGPT 185
Query 141 HGSIIQLQGDQRQAVKEFL 159
I +QGD + EF+
Sbjct 186 EKEQIDVQGDIIYDIVEFI 204
> Hs18571842
Length=107
Score = 32.0 bits (71), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 144 IIQLQGDQRQAVKEFLEREGICGAEQLRIHG 174
+IQLQGDQ + + +FL G+ QL +HG
Sbjct 76 VIQLQGDQHKNMCQFLLEIGMAKDNQLNVHG 106
> YJR014w
Length=198
Score = 29.6 bits (65), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 16/66 (24%), Positives = 32/66 (48%), Gaps = 1/66 (1%)
Query 95 IRNQQRNGRKSVTTVQGLPK-AFDLKKMVRALKKEFSCNGTVIEDPEHGSIIQLQGDQRQ 153
I+ + R RK + + GL D+KK+ + F+ +V ++ E + +QGD
Sbjct 101 IKREARTKRKFIVAISGLEVFDIDMKKLAKTFASRFATGCSVSKNAEKKEEVVIQGDVMD 160
Query 154 AVKEFL 159
V+ ++
Sbjct 161 EVETYI 166
> At5g25930
Length=1005
Score = 28.1 bits (61), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 15/56 (26%), Positives = 30/56 (53%), Gaps = 5/56 (8%)
Query 114 KAFDLKKMVRALKKEFSCNGTVIEDPEHGSIIQL-----QGDQRQAVKEFLEREGI 164
+ +D KK+ + L+KEF ++ H +I++L + D + V E+LE+ +
Sbjct 715 RIWDSKKLDQKLEKEFIAEVEILGTIRHSNIVKLLCCISREDSKLLVYEYLEKRSL 770
Lambda K H
0.324 0.140 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2671071884
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40