bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3160_orf2
Length=307
Score E
Sequences producing significant alignments: (Bits) Value
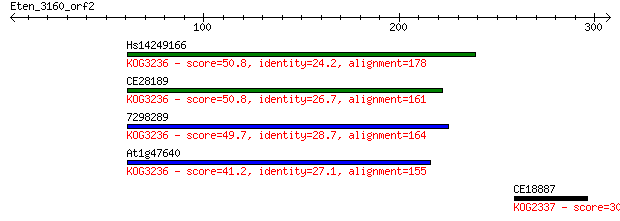
Hs14249166 50.8 3e-06
CE28189 50.8 4e-06
7298289 49.7 9e-06
At1g47640 41.2 0.003
CE18887 30.0 6.6
> Hs14249166
Length=224
Score = 50.8 bits (120), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 43/178 (24%), Positives = 79/178 (44%), Gaps = 3/178 (1%)
Query 61 MTVLHGAACAAFCFGPIAAIYKATNLSDQTGTKKLLALATAFYFVNLFGKSLIVATLLPS 120
MT+ H C A + P YK + LS+ K + + FV L K L +AT P
Sbjct 1 MTLFHFGNCFALAYFPYFITYKCSGLSEYNAFWKCVQAGVTYLFVQL-CKMLFLATFFP- 58
Query 121 TCESSFFCSVIIDLVTVVMELXSIRYIXHSKKALAYSSETRLLCIGLGWSLGQVLCSNGH 180
T E + I + + +++ + + A E +++ LGW+ +++ S
Sbjct 59 TWEGGIY-DFIGEFMKASVDVADLIGLNLVMSRNAGKGEYKIMVAALGWATAELIMSRCI 117
Query 181 LLLRAIGGNSFHWEGVYQAMQSSLSPISYLSITALMFLSTRRHLPMTSRAGLVFLASL 238
L G F W+ + ++ S++S + Y+ +A +++ TR L T R ++ L L
Sbjct 118 PLWVGARGIEFDWKYIQMSIDSNISLVHYIVASAQVWMITRYDLYHTFRPAVLLLMFL 175
> CE28189
Length=225
Score = 50.8 bits (120), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 43/167 (25%), Positives = 69/167 (41%), Gaps = 14/167 (8%)
Query 61 MTVLHGAACAAFCFGPIAAIYKATNLSDQTGTKKLLALATAFYFVNLFGKSLIVATLLPS 120
M+ H C A F P +YK + +++ + K A A+ Y + K LI+AT P+
Sbjct 1 MSFFHFINCFALAFAPYFIVYKYSGINEYSSIWKC-ATASGGYLLTQLAKLLIIATFFPA 59
Query 121 TCESSF-----FCSVIIDLVTVV-MELXSIRYIXHSKKALAYSSETRLLCIGLGWSLGQV 174
F F D++ V+ + L + LA E R + GLGW
Sbjct 60 LDSEGFSIVPEFLKSSADIIDVIGLHLLMTNF-------LAGKGEVRFVVGGLGWGFAHS 112
Query 175 LCSNGHLLLRAIGGNSFHWEGVYQAMQSSLSPISYLSITALMFLSTR 221
+ LL G +F W V ++ SS + +S+ L ++ TR
Sbjct 113 VAHRLVLLWVGARGTAFTWRWVQTSLDSSADLLVIVSLACLTWMITR 159
> 7298289
Length=227
Score = 49.7 bits (117), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 47/175 (26%), Positives = 76/175 (43%), Gaps = 23/175 (13%)
Query 61 MTVLHGAACAAFCFGPIAAIYKATNLSDQTGTKKLLALATAFYFVNLFGKSLIVATLL-- 118
MT+ H C A P YK + LS+ K + + F L K L++AT
Sbjct 1 MTLYHFGNCVAL-LTPYYFTYKYSGLSEYGAFWKCVQAGGIYIFTQLV-KMLVLATFFYS 58
Query 119 ---PSTCESSFFCSV------IIDLVTVVMELXSIRYIXHSKKALAYSSETRLLCIGLGW 169
S+ E +FF + I DL+ + L I HSK L+ GLGW
Sbjct 59 DAPSSSGEFNFFAEILRCSMDIADLLGFALILSRIPGKGHSK----------LITAGLGW 108
Query 170 SLGQVLCSNGHLLLRAIGGNSFHWEGVYQAMQSSLSPISYLSITALMFLSTRRHL 224
+ +V+ S G +L G F W + + ++S++ + +++ L++L TR L
Sbjct 109 ATAEVILSRGIMLWVGARGTEFSWIYILKCLESNVLLVQHITTATLIWLFTRHDL 163
> At1g47640
Length=228
Score = 41.2 bits (95), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 42/163 (25%), Positives = 64/163 (39%), Gaps = 16/163 (9%)
Query 61 MTVLHGAACAAFCFGPIAAIYKATNLS--DQTGTKKLLALATAFYFVNLFGKSLIVATLL 118
MT+ H CA FGP A Y AT LS D GT A Y K + +AT L
Sbjct 1 MTLFHFFNCAILTFGPHAVYYSATPLSEYDTLGTS---VKAAVVYLATALVKLVCLATFL 57
Query 119 PSTCESSFF------CSVIIDLVTVVMELXSIRYIXHSKKALAYSSETRLLCIGLGWSLG 172
E+ F +I + V ++ + H S + +GLGW+
Sbjct 58 -QVSETEVFDPYQEALKAMIGFIDVAGLYYALAQLTHRN----ISQNHKFQAVGLGWAFA 112
Query 173 QVLCSNGHLLLRAIGGNSFHWEGVYQAMQSSLSPISYLSITAL 215
+ L G F W+ V Q ++++ + + +S+ AL
Sbjct 113 DSVLHRLAPLWVGARGLEFTWDYVLQGLEANANLVFTISLAAL 155
> CE18887
Length=647
Score = 30.0 bits (66), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 24/42 (57%), Gaps = 5/42 (11%)
Query 259 FLAEDITPACWLSLVLHA-----AVSISVGITTYLTLRAGHG 295
FLA D A WL VL + A+S+++G TY+ +R G G
Sbjct 438 FLALDSREARWLPTVLASRHKKIAISVAIGFDTYVIIRHGIG 479
Lambda K H
0.327 0.137 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7025720074
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40