bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3137_orf1
Length=180
Score E
Sequences producing significant alignments: (Bits) Value
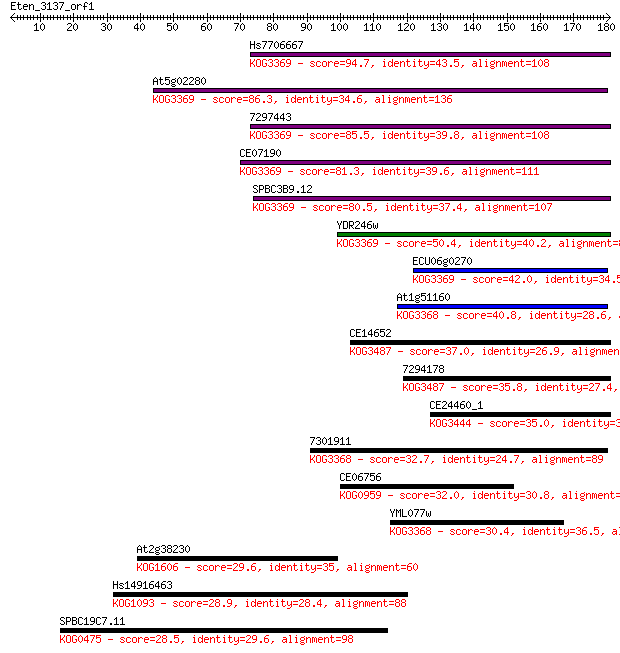
Hs7706667 94.7 8e-20
At5g02280 86.3 3e-17
7297443 85.5 5e-17
CE07190 81.3 9e-16
SPBC3B9.12 80.5 2e-15
YDR246w 50.4 2e-06
ECU06g0270 42.0 7e-04
At1g51160 40.8 0.001
CE14652 37.0 0.019
7294178 35.8 0.043
CE24460_1 35.0 0.074
7301911 32.7 0.37
CE06756 32.0 0.74
YML077w 30.4 1.9
At2g38230 29.6 3.5
Hs14916463 28.9 6.2
SPBC19C7.11 28.5 6.7
> Hs7706667
Length=219
Score = 94.7 bits (234), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 47/109 (43%), Positives = 70/109 (64%), Gaps = 14/109 (12%)
Query 73 ANDAIRQASTFHGLSEIAAQLSPLSCFSSSDSSSIQPRGICLIETPQFNVQCLETLTGLR 132
+N+ + AS FH L I +QLSP S GI ++ET F + C +TLTG++
Sbjct 105 SNEKLMLASMFHSLFAIGSQLSPEQGSS----------GIEMLETDTFKLHCYQTLTGIK 154
Query 133 FVLVADPTLSRQS-VEQCLRRVYVAYADYGLKNPFYDIDMPLRCPLFEQ 180
FV++ADP RQ+ ++ LR++Y Y+D+ LKNPFY ++MP+RC LF+Q
Sbjct 155 FVVLADP---RQAGIDSLLRKIYEIYSDFALKNPFYSLEMPIRCELFDQ 200
> At5g02280
Length=141
Score = 86.3 bits (212), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 47/136 (34%), Positives = 76/136 (55%), Gaps = 12/136 (8%)
Query 44 LAFSFCLRLFSCSGIYVALQNFKEGISVKANDAIRQASTFHGLSEIAAQLSPLSCFSSSD 103
+A + L + + SG + ++ + ND++R AS +H + I+ QLSP++ S
Sbjct 1 MAAIYSLYIINKSGGLIFYKDCGTKGRMDTNDSLRVASLWHSMHAISQQLSPVNGCS--- 57
Query 104 SSSIQPRGICLIETPQFNVQCLETLTGLRFVLVADPTLSRQSVEQCLRRVYVAYADYGLK 163
GI L+E F++ C ++L G +F +V +P +E LR +Y Y DY LK
Sbjct 58 -------GIELLEADTFDLHCFQSLPGTKFFVVCEP--GTPHMESLLRYIYELYTDYVLK 108
Query 164 NPFYDIDMPLRCPLFE 179
NPFY+I+MP+RC LF+
Sbjct 109 NPFYEIEMPIRCELFD 124
> 7297443
Length=219
Score = 85.5 bits (210), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 43/110 (39%), Positives = 68/110 (61%), Gaps = 16/110 (14%)
Query 73 ANDAIRQASTFHGLSEIAAQLSPLSCFSSSDSSSIQPR--GICLIETPQFNVQCLETLTG 130
N+ I AS F+ L IA+QLSP +P+ GI ++E F + C +TLTG
Sbjct 105 TNEKIFLASMFYPLFAIASQLSP------------EPKSSGIEILEADTFTLHCFQTLTG 152
Query 131 LRFVLVADPTLSRQSVEQCLRRVYVAYADYGLKNPFYDIDMPLRCPLFEQ 180
++F+++++ L+ ++ LR+VY Y+DY LKNPFY ++MP+RC LF+
Sbjct 153 IKFIIISETGLN--GIDLLLRKVYELYSDYVLKNPFYSLEMPIRCELFDN 200
> CE07190
Length=224
Score = 81.3 bits (199), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 44/111 (39%), Positives = 65/111 (58%), Gaps = 11/111 (9%)
Query 70 SVKANDAIRQASTFHGLSEIAAQLSPLSCFSSSDSSSIQPRGICLIETPQFNVQCLETLT 129
+V N+ I +S FH L IA QLSP C SS G+ ++ET QF + CL++ T
Sbjct 107 TVSTNEKIILSSMFHSLFTIAVQLSP--CQKSS--------GVEVLETTQFKLFCLQSRT 156
Query 130 GLRFVLVADPTLSRQSVEQCLRRVYVAYADYGLKNPFYDIDMPLRCPLFEQ 180
G++FV++ S + + L ++Y Y D+ LKNPFY IDMP+R F++
Sbjct 157 GVKFVVITSAA-SNIAADSLLSKMYELYTDFALKNPFYSIDMPIRAQKFDE 206
> SPBC3B9.12
Length=132
Score = 80.5 bits (197), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 40/107 (37%), Positives = 63/107 (58%), Gaps = 12/107 (11%)
Query 74 NDAIRQASTFHGLSEIAAQLSPLSCFSSSDSSSIQPRGICLIETPQFNVQCLETLTGLRF 133
N+ + A T HG+ I+ Q+SPL S GI L+E FN+ L+T TG++F
Sbjct 29 NEYLVLAGTIHGVHAISTQISPLPGSS----------GIQLLEAGTFNMHILQTHTGMKF 78
Query 134 VLVADPTLSRQSVEQCLRRVYVAYADYGLKNPFYDIDMPLRCPLFEQ 180
VL + + ++ L++ Y Y+DY LKNPFY ++MP++C LF++
Sbjct 79 VLFTEKKTTNARLQ--LQKFYELYSDYVLKNPFYTLEMPIKCQLFDE 123
> YDR246w
Length=219
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/104 (31%), Positives = 54/104 (51%), Gaps = 27/104 (25%)
Query 99 FSSSDSSSIQPRGICLIETPQFNVQCLETLTGLRFVLVA-------DPT----------- 140
F++ + S + R +C T QF + +TLTGL+FV ++ PT
Sbjct 111 FTNWNKSGL--RQLC---TDQFTMFIYQTLTGLKFVAISSSVMPQRQPTIATTDKPDRPK 165
Query 141 ----LSRQSVEQCLRRVYVAYADYGLKNPFYDIDMPLRCPLFEQ 180
L+ Q + LR+VY Y+DY +K+P Y ++MP+R LF++
Sbjct 166 STSNLAIQIADNFLRKVYCLYSDYVMKDPSYSMEMPIRSNLFDE 209
> ECU06g0270
Length=123
Score = 42.0 bits (97), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 30/58 (51%), Gaps = 5/58 (8%)
Query 122 VQCLETLTGLRFVLVADPTLSRQSVEQCLRRVYVAYADYGLKNPFYDIDMPLRCPLFE 179
+ T TG FV VAD + V+ R+Y Y Y +NPF+ +MP++C F+
Sbjct 64 ITIFRTTTGTSFVFVAD-----RPVDALFERIYSHYCQYVTRNPFHSPEMPIQCSKFK 116
> At1g51160
Length=169
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/63 (28%), Positives = 33/63 (52%), Gaps = 2/63 (3%)
Query 117 TPQFNVQCLETLTGLRFVLVADPTLSRQSVEQCLRRVYVAYADYGLKNPFYDIDMPLRCP 176
T + + +ET +G++ +LV P + + L+ +Y Y +Y +KNP Y P++
Sbjct 98 TNTYKLSFMETPSGIKIILVTHPKTG--DLRESLKYIYSLYVEYVVKNPIYSPGSPIKSE 155
Query 177 LFE 179
LF
Sbjct 156 LFN 158
> CE14652
Length=141
Score = 37.0 bits (84), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 40/81 (49%), Gaps = 5/81 (6%)
Query 103 DSSSIQPRGICLIETPQFN---VQCLETLTGLRFVLVADPTLSRQSVEQCLRRVYVAYAD 159
D ++ + L +FN V T + +RF+++ + + ++Q + +Y Y
Sbjct 52 DEHALTTSQMYLKMVDKFNEWYVSAFVTASRIRFIMLH-THRADEGIKQFFQEMYETYIK 110
Query 160 YGLKNPFYDIDMPLRCPLFEQ 180
+ + NPFY+ID + P FEQ
Sbjct 111 HAM-NPFYEIDDVIESPAFEQ 130
> 7294178
Length=158
Score = 35.8 bits (81), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 33/62 (53%), Gaps = 2/62 (3%)
Query 119 QFNVQCLETLTGLRFVLVADPTLSRQSVEQCLRRVYVAYADYGLKNPFYDIDMPLRCPLF 178
Q+ V T + +RF++V D + + ++ +Y Y + N FY I+ P++ P+F
Sbjct 87 QWFVSAFITASQIRFIIVHDNK-NDEGIKNFFNEMYDTYIKNSM-NAFYRINTPIKSPMF 144
Query 179 EQ 180
E+
Sbjct 145 EK 146
> CE24460_1
Length=115
Score = 35.0 bits (79), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 30/58 (51%), Gaps = 5/58 (8%)
Query 127 TLTGLRFVLVADPT----LSRQSVEQCLRRVYVAYADYGLKNPFYDIDMPLRCPLFEQ 180
T TG+R +LV D T L Q + +R + Y + + NPFY+I P++ ++
Sbjct 38 TNTGVRMILVLDATSAASLKDQEIRLIFKRFHGHYCN-TISNPFYEIGTPMQSKWLDE 94
> 7301911
Length=145
Score = 32.7 bits (73), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 22/90 (24%), Positives = 44/90 (48%), Gaps = 3/90 (3%)
Query 91 AQLSPLSCFSSSDSSSIQPRGICLIETPQFNVQCLETLTGLRFVLVADPTLSRQSVEQCL 150
L + F S S G +T ++ + LET +GL+FVL D T +V++ L
Sbjct 40 GMLFSIKSFVSKISPHDPKEGFLYYKTNRYALHYLETPSGLKFVLNTDTTAI--NVKELL 97
Query 151 RRVYV-AYADYGLKNPFYDIDMPLRCPLFE 179
+++Y + ++ +++P + + LF+
Sbjct 98 QQLYAKVWVEFVVRDPLWTPGTVVTSELFQ 127
> CE06756
Length=845
Score = 32.0 bits (71), Expect = 0.74, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 30/54 (55%), Gaps = 2/54 (3%)
Query 100 SSSDSSSIQPRGICLIETPQFNVQC--LETLTGLRFVLVADPTLSRQSVEQCLR 151
+ +D + + R +++ PQ +C LE GLR +LV+DPT + +V ++
Sbjct 2 TEADRNIVLKRYNSIVKGPQDERECRGLELTNGLRVLLVSDPTTDKSAVSLAVK 55
> YML077w
Length=159
Score = 30.4 bits (67), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 29/56 (51%), Gaps = 6/56 (10%)
Query 115 IETPQFNVQCLETLTGLRFVLVADPTLSRQSVEQCLR----RVYVAYADYGLKNPF 166
I T ++ V T +GL FVL++D +QS Q L+ +YV Y L +P+
Sbjct 70 ISTGKYRVHTYCTASGLWFVLLSD--FKQQSYTQVLQYIYSHIYVKYVSNNLLSPY 123
> At2g38230
Length=309
Score = 29.6 bits (65), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 29/60 (48%), Gaps = 3/60 (5%)
Query 39 GHIGTLAFSFCLRLFSCSGIYVALQNFKEGISVKANDAIRQASTFHGLSEIAAQLSPLSC 98
G + T A + + C G++V FK G VK AI QA T + AA L+ +SC
Sbjct 230 GGVATPADAALMMQLGCDGVFVGSGVFKSGDPVKRAKAIVQAVTNY---RDAAVLAEVSC 286
> Hs14916463
Length=822
Score = 28.9 bits (63), Expect = 6.2, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 39/94 (41%), Gaps = 8/94 (8%)
Query 32 SSRCLLRGHI-GTLAFSFCLRLFS----CSGIYVA-LQNFKEGISVKANDAIRQASTFHG 85
+S +RGHI G++ F + G Y A LQNFK + V + + F
Sbjct 730 NSEDFIRGHISGSINIPFSAAFTAEGELTQGPYTAMLQNFKGKVIVIVGHVAKHTAEF-- 787
Query 86 LSEIAAQLSPLSCFSSSDSSSIQPRGICLIETPQ 119
+ + P C + I+P G+ I +PQ
Sbjct 788 AAHLVKMKYPRICILDGGINKIKPTGLLTIPSPQ 821
> SPBC19C7.11
Length=812
Score = 28.5 bits (62), Expect = 6.7, Method: Composition-based stats.
Identities = 29/111 (26%), Positives = 51/111 (45%), Gaps = 25/111 (22%)
Query 16 SIAAVFVVAAACVV-----------LCSSRCLLRGHIGTLAFSFCLRLFSCSGIYVALQN 64
+ A +FV+ AA +V + +C++ G + SF + L C G+ +A+ +
Sbjct 185 AFALLFVLCAAIMVRDVAPLAAGSGISEIKCIISGFLRDSFLSFRVMLVKCVGLPLAIAS 244
Query 65 FKEGISV-KANDAIRQASTF-HGLSEIAAQLSPLSCFSSSDSSSIQPRGIC 113
G+SV K ++ A+T H +S+I F + SI+ R IC
Sbjct 245 ---GLSVGKEGPSVHLATTIGHNISKI---------FKYAREGSIRYRDIC 283
Lambda K H
0.328 0.138 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2806646388
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40