bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3136_orf1
Length=229
Score E
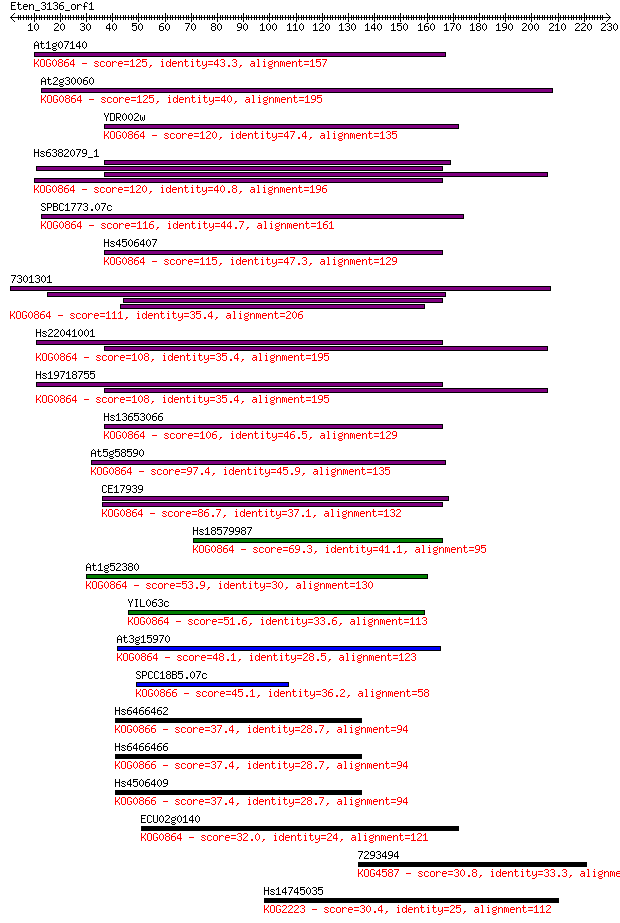
Sequences producing significant alignments: (Bits) Value
At1g07140 125 8e-29
At2g30060 125 1e-28
YDR002w 120 2e-27
Hs6382079_1 120 3e-27
SPBC1773.07c 116 4e-26
Hs4506407 115 1e-25
7301301 111 1e-24
Hs22041001 108 7e-24
Hs19718755 108 1e-23
Hs13653066 106 4e-23
At5g58590 97.4 2e-20
CE17939 86.7 4e-17
Hs18579987 69.3 7e-12
At1g52380 53.9 2e-07
YIL063c 51.6 1e-06
At3g15970 48.1 2e-05
SPCC18B5.07c 45.1 1e-04
Hs6466462 37.4 0.023
Hs6466466 37.4 0.028
Hs4506409 37.4 0.028
ECU02g0140 32.0 0.98
7293494 30.8 2.3
Hs14745035 30.4 2.7
> At1g07140
Length=228
Score = 125 bits (314), Expect = 8e-29, Method: Compositional matrix adjust.
Identities = 68/158 (43%), Positives = 91/158 (57%), Gaps = 4/158 (2%)
Query 10 TQAPAQTAEDDEVANLEQEVTEGNWTRPEVEVHEVKVETGEESEDTFWKCRSKLYRWAA- 68
T P D+E A ++ G P V + EV V TGEE ED +SKLYR+
Sbjct 3 TNEPEHEHRDEEEAGANEDEDTGAQVAPIVRLEEVAVTTGEEDEDAVLDLKSKLYRFDKD 62
Query 69 GGEWKERGLGEAKLLQHRETKKIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIW 128
+WKERG G K L+H+ T KIR ++RQ KTLKI ANH+V + + +V +EK
Sbjct 63 ANQWKERGAGTVKFLKHKNTGKIRLVMRQSKTLKICANHFVKSG---MSVQEHVGNEKSC 119
Query 129 VWTVMDFAEGELKNEQFALKFGQVEQAKEFKEKFEEAA 166
VW DFA+GELK+E F ++F +E K F +KF+E A
Sbjct 120 VWHARDFADGELKDELFCIRFASIENCKTFMQKFKEVA 157
> At2g30060
Length=217
Score = 125 bits (313), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 78/201 (38%), Positives = 109/201 (54%), Gaps = 9/201 (4%)
Query 13 PAQTAEDDEVANLEQEVTEGNWTRPEVEVHEVKVETGEESEDTFWKCRSKLYRWAA-GGE 71
P + D+E ++ G P V + EV V TGEE EDT +SKLYR+ G +
Sbjct 8 PERENRDEEETGANEDEDTGAQVAPIVRLEEVAVTTGEEDEDTILDLKSKLYRFDKDGSQ 67
Query 72 WKERGLGEAKLLQHRETKKIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWT 131
WKERG G K L+HR + KIR ++RQ KTLKI ANH V + + + ++K VW
Sbjct 68 WKERGAGTVKFLKHRVSGKIRLVMRQSKTLKICANHLVGSG---MSVQEHAGNDKSCVWH 124
Query 132 VMDFAEGELKNEQFALKFGQVEQAKEFKEKFEEAAAINSKLFGIGDSTK-----EGEKTE 186
DF++GELK+E F ++F VE K F +KF+E A + D++ E E
Sbjct 125 ARDFSDGELKDELFCIRFASVENCKAFMQKFKEVAESEEEKEESKDASDTAGLLEKLTVE 184
Query 187 EKEAEKSTESKDDETKETEGA 207
EKE+EK K +E K++E
Sbjct 185 EKESEKKPVEKAEENKKSEAV 205
> YDR002w
Length=201
Score = 120 bits (302), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 64/137 (46%), Positives = 93/137 (67%), Gaps = 5/137 (3%)
Query 37 PEVEVHEVKVETGEESEDTFWKCRSKLYRWAAGG-EWKERGLGEAKLLQHRETKKIRFLL 95
P V + +V V+T EE E+ +K R+KL+R+ A EWKERG G+ K L++++T K+R L+
Sbjct 67 PVVHLEKVDVKTMEEDEEVLYKVRAKLFRFDADAKEWKERGTGDCKFLKNKKTNKVRILM 126
Query 96 RQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVM-DFAEGELKNEQFALKFGQVEQ 154
R++KTLKI ANH ++A + L PNV S++ WV+ D AEGE + FA++FG E
Sbjct 127 RRDKTLKICANH-IIAPEYT--LKPNVGSDRSWVYACTADIAEGEAEAFTFAIRFGSKEN 183
Query 155 AKEFKEKFEEAAAINSK 171
A +FKE+FE+A IN K
Sbjct 184 ADKFKEEFEKAQEINKK 200
> Hs6382079_1
Length=3065
Score = 120 bits (300), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 62/134 (46%), Positives = 93/134 (69%), Gaps = 5/134 (3%)
Query 37 PEVEVHE-VKVETGEESEDTFWKCRSKLYRWAA-GGEWKERGLGEAKLLQHRETKKIRFL 94
P V + + ++V+TGEE E+ F+ R+KL+R+ EWKERG+G K+L+H+ + KIR L
Sbjct 1174 PVVPLPDKIEVKTGEEDEEEFFCNRAKLFRFDVESKEWKERGIGNVKILRHKTSGKIRLL 1233
Query 95 LRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAEGELKNEQFALKFGQVEQ 154
+R+E+ LKI ANHY ++ D+ KLTPN S++ +VW +D+A+ K EQ A++F E+
Sbjct 1234 MRREQVLKICANHY-ISPDM--KLTPNAGSDRSFVWHALDYADELPKPEQLAIRFKTPEE 1290
Query 155 AKEFKEKFEEAAAI 168
A FK KFEEA +I
Sbjct 1291 AALFKCKFEEAQSI 1304
Score = 108 bits (269), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 57/163 (34%), Positives = 100/163 (61%), Gaps = 17/163 (10%)
Query 11 QAPAQTAEDDEVANLEQEVTEGNWTRPEVEVHE-VKVETGEESEDTFWKCRSKLYRWAAG 69
Q+ D+E ++E +G + P V + + V+V +GEE+E + R+KLYR+
Sbjct 2286 QSGTSVGTDEESDVTQEEERDGQYFEPVVPLPDLVEVSSGEENEQVVFSHRAKLYRYDKD 2345
Query 70 -GEWKERGLGEAKLLQHRETKKIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVS----- 123
G+WKERG+G+ K+LQ+ + K++R ++R+++ LK+ ANH ++TP+++
Sbjct 2346 VGQWKERGIGDIKILQNYDNKQVRIVMRRDQVLKLCANH---------RITPDMTLQNMK 2396
Query 124 -SEKIWVWTVMDFAEGELKNEQFALKFGQVEQAKEFKEKFEEA 165
+E++W+WT DFA+GE K E A++F + A FK+ F+EA
Sbjct 2397 GTERVWLWTACDFADGERKVEHLAVRFKLQDVADSFKKIFDEA 2439
Score = 107 bits (268), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 69/182 (37%), Positives = 104/182 (57%), Gaps = 16/182 (8%)
Query 37 PEVEVHE-VKVETGEESEDTFWKCRSKLYRWAA-GGEWKERGLGEAKLLQHRETKKIRFL 94
P V++ E V++ TGEE E + R KL+R+ A +WKERGLG K+L++ K+R L
Sbjct 2015 PVVQMPEKVELVTGEEDEKVLYSQRVKLFRFDAEVSQWKERGLGNLKILKNEVNGKLRML 2074
Query 95 LRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAEGELKNEQFALKFGQVEQ 154
+R+E+ LK+ ANH++ T L P S++ W+W DF++G+ K EQ A KF E
Sbjct 2075 MRREQVLKVCANHWITTT---MNLKPLSGSDRAWMWLASDFSDGDAKLEQLAAKFKTPEL 2131
Query 155 AKEFKEKFEEAAAI--------NSKLFGIGDSTKEGEKTEEKEA---EKSTESKDDETKE 203
A+EFK+KFEE + KL G + K ++ EE ++ + T +D+TK
Sbjct 2132 AEEFKQKFEECQRLLLDIPLQTPHKLVDTGRAAKLIQRAEEMKSGLKDFKTFLTNDQTKV 2191
Query 204 TE 205
TE
Sbjct 2192 TE 2193
Score = 103 bits (257), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 61/157 (38%), Positives = 90/157 (57%), Gaps = 4/157 (2%)
Query 10 TQAPAQTAEDDEVANLEQEVTEGNWTRPEVEVHEVKVETGEESEDTFWKCRSKLYRWAAG 69
TQ+ + ED++ ++ E E P V + EV+V++GEE E+ +K R+KLYRW
Sbjct 2887 TQSAGKVGEDEDGSDEEVVHNEDIHFEPIVSLPEVEVKSGEEDEEILFKERAKLYRWDRD 2946
Query 70 -GEWKERGLGEAKLLQHRETKKIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIW 128
+WKERG+G+ K+L H R L+R+++ K+ ANH + T +L P S
Sbjct 2947 VSQWKERGVGDIKILWHTMKNYYRILMRRDQVFKVCANHVITKT---MELKPLNVSNNAL 3003
Query 129 VWTVMDFAEGELKNEQFALKFGQVEQAKEFKEKFEEA 165
VWT D+A+GE K EQ A++F E A FK+ FEE
Sbjct 3004 VWTASDYADGEAKVEQLAVRFKTKEVADCFKKTFEEC 3040
> SPBC1773.07c
Length=215
Score = 116 bits (290), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 72/163 (44%), Positives = 101/163 (61%), Gaps = 11/163 (6%)
Query 13 PAQTAEDDEVANLEQEVTEGNWTRPEVEVHEVKVETGEESEDTFWKCRSKLYRW-AAGGE 71
PA+ ED E A+ E P V++ V+ +T EE E +K R+KL+R+ A E
Sbjct 59 PAEGDEDAEPASPEVHF------EPIVKLSAVETKTNEEEETVEFKMRAKLFRFDKAASE 112
Query 72 WKERGLGEAKLLQHRETKKIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWT 131
WKERG G+A+LL+H+ET K R ++R++KTLK+ ANH ++ KLTPNV S++ WVWT
Sbjct 113 WKERGTGDARLLKHKETGKTRLVMRRDKTLKVCANHLLMPE---MKLTPNVGSDRSWVWT 169
Query 132 V-MDFAEGELKNEQFALKFGQVEQAKEFKEKFEEAAAINSKLF 173
V D +EGE E FA++F E A FKE FE+ N+K+
Sbjct 170 VAADVSEGEPTAETFAIRFANSENANLFKENFEKYQEENAKIL 212
> Hs4506407
Length=201
Score = 115 bits (287), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 61/133 (45%), Positives = 86/133 (64%), Gaps = 7/133 (5%)
Query 37 PEVEVHEVKVETGEESEDTFWKCRSKLYRWAAGG---EWKERGLGEAKLLQHRETKKIRF 93
P V + E +++T EE E+ +K R+KL+R+A+ EWKERG G+ KLL+H+E IR
Sbjct 29 PIVSLPEQEIKTLEEDEEELFKMRAKLFRFASENDLPEWKERGTGDVKLLKHKEKGAIRL 88
Query 94 LLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVW-TVMDFAEGELKNEQFALKFGQV 152
L+R++KTLKI ANHY+ +L PN S++ WVW T DFA+ K E A++F
Sbjct 89 LMRRDKTLKICANHYITP---MMELKPNAGSDRAWVWNTHADFADECPKPELLAIRFLNA 145
Query 153 EQAKEFKEKFEEA 165
E A++FK KFEE
Sbjct 146 ENAQKFKTKFEEC 158
> 7301301
Length=2659
Score = 111 bits (277), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 73/210 (34%), Positives = 112/210 (53%), Gaps = 12/210 (5%)
Query 1 YKMADKETETQAPAQTAEDDEVANLEQEVTE---GNWTRPEVEVHE-VKVETGEESEDTF 56
+ M + + Q P TA ++ N QEV E + P + + + + V+TGEE E+
Sbjct 1947 FSMTKPKPDQQQPNSTAAKEDEDNDSQEVEEEENNTYFSPVIPLPDKIDVKTGEEDEELL 2006
Query 57 WKCRSKLYRWAAGGEWKERGLGEAKLLQHRETKKIRFLLRQEKTLKIVANHYVVATDVYC 116
+ ++KLYR +WKERGLG+ K+L+HR+TKK+R ++R+E+ KI NH + VY
Sbjct 2007 YVHKAKLYRLNES-DWKERGLGDVKILRHRQTKKLRVVMRREQVFKICLNHVLNENVVYR 2065
Query 117 KLTPNVSSEKIWVWTVMDFAEGELKNEQFALKFGQVEQAKEFKEKFEEAAAINSKLFGIG 176
+ T E W++ V DF+EGE E+F L+F E A+ F E + A+N I
Sbjct 2066 EKT-----ETSWMFAVHDFSEGESVLERFTLRFKNKEVAQGFMEAIKN--ALNETAKPIE 2118
Query 177 DSTKEGEKTEEKEAEKSTESKDDETKETEG 206
DS G ++ EA K ++ D K G
Sbjct 2119 DSPVVGSVSQSTEANKPSQKNDGAAKSRGG 2148
Score = 83.6 bits (205), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 53/158 (33%), Positives = 87/158 (55%), Gaps = 8/158 (5%)
Query 15 QTAEDDEVANLEQEVTEGNW-TRPEVEV-----HEVKVETGEESEDTFWKCRSKLYRWAA 68
Q+ D L Q+ + ++ RP+ + EV+V TGEE ED + R+KL+R+
Sbjct 1249 QSGATDPNKTLPQDTSADDYDPRPDFKPIIPLPDEVEVRTGEEGEDIKFTSRAKLFRYV- 1307
Query 69 GGEWKERGLGEAKLLQHRETKKIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIW 128
EWKERG G K+L + T R L+R+++T K+ ANH + A D+ + +K
Sbjct 1308 DKEWKERGTGVIKILCDKATGVSRVLMRRDQTHKVCANHTITA-DITINVANQDKDKKSL 1366
Query 129 VWTVMDFAEGELKNEQFALKFGQVEQAKEFKEKFEEAA 166
+W DFA+ ++ E+F ++F E A+EF+ F +A+
Sbjct 1367 LWAANDFADEQVTLERFLVRFKTGELAEEFRVAFTKAS 1404
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 43/124 (34%), Positives = 69/124 (55%), Gaps = 5/124 (4%)
Query 44 VKVETGEESEDTFWKCRSKLYRWAA-GGEWKERGLGEAKLLQHR-ETKKIRFLLRQEKTL 101
V+V TGEE+ED ++ R+KL RW EWKERGLG KLL+ R + K+R L+R+E+
Sbjct 1580 VEVVTGEENEDVLFEHRAKLLRWDKEANEWKERGLGNMKLLRDRTDPNKVRLLMRREQVH 1639
Query 102 KIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAEGELKNEQFALKFGQVEQAKEFKEK 161
K+ N ++ K T + + W D+++ EL A++F + ++F E
Sbjct 1640 KLCCNQRLLPE---TKFTYATNCKAAVTWGAQDYSDEELTTALLAVRFKSQDICQQFLEA 1696
Query 162 FEEA 165
++A
Sbjct 1697 VQKA 1700
Score = 70.1 bits (170), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 44/124 (35%), Positives = 72/124 (58%), Gaps = 11/124 (8%)
Query 43 EVKVETGEESEDTFWKCRSKLYRWAA-GGEWKERGLGEAKLLQHRETKKIRFLLRQEKTL 101
E+ V TGEE+E + R+KLYR+ A +WKERG+GE K+L+H E + R ++RQE+
Sbjct 2507 EIVVTTGEENETKLFGERAKLYRYDAESKQWKERGVGEIKVLEHPELQTFRLIMRQEQIH 2566
Query 102 KIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDF---AEGELKN----EQFALKFGQVEQ 154
K+V N + A+ ++ + K ++W ++ AEG++ E+ A +F + E
Sbjct 2567 KLVLNMNISAS---LQMDYMNAQMKSFLWAGYNYAVDAEGKVDTEGVLERLACRFAKEEI 2623
Query 155 AKEF 158
A EF
Sbjct 2624 ASEF 2627
> Hs22041001
Length=1021
Score = 108 bits (271), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 57/163 (34%), Positives = 100/163 (61%), Gaps = 17/163 (10%)
Query 11 QAPAQTAEDDEVANLEQEVTEGNWTRPEVEVHE-VKVETGEESEDTFWKCRSKLYRWAAG 69
Q+ D+E ++E +G + P V + + V+V +GEE+E + R+++YR+
Sbjct 566 QSGTSVGTDEESVVTQEEERDGQYFEPVVPLPDLVEVSSGEENEQVVFSHRAEIYRYDKD 625
Query 70 -GEWKERGLGEAKLLQHRETKKIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVS----- 123
G+WKERG+G+ K+LQ+ + K++R ++R+++ LK+ ANH ++TP++S
Sbjct 626 VGQWKERGIGDIKILQNYDNKQVRIVMRRDQVLKLCANH---------RITPDMSLQNMK 676
Query 124 -SEKIWVWTVMDFAEGELKNEQFALKFGQVEQAKEFKEKFEEA 165
+E++WVWT DFA+GE K E A++F + A FK+ F+EA
Sbjct 677 GTERVWVWTACDFADGERKVEHLAVRFKLQDVADSFKKIFDEA 719
Score = 103 bits (256), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 67/182 (36%), Positives = 104/182 (57%), Gaps = 16/182 (8%)
Query 37 PEVEVHE-VKVETGEESEDTFWKCRSKLYRWAA-GGEWKERGLGEAKLLQHRETKKIRFL 94
P V++ E V++ TGEE E + KL+R+ A +WKERGLG K+L++ K+R L
Sbjct 295 PVVQMPEKVELVTGEEGEKVLYSQGVKLFRFDAEVRQWKERGLGNLKILKNEVNGKLRML 354
Query 95 LRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAEGELKNEQFALKFGQVEQ 154
+R+E+ LK+ ANH++ T L P S++ W+W+ DF++G+ K E+ A KF E
Sbjct 355 MRREQVLKVCANHWITTT---MNLKPLSGSDRAWMWSASDFSDGDAKLERLAAKFKTPEL 411
Query 155 AKEFKEKFEEAAAI--------NSKLFGIGDSTKEGEKTEEKEA---EKSTESKDDETKE 203
A+EFK+KFEE + KL G + K ++ EE ++ + T +D+TK
Sbjct 412 AEEFKQKFEECQRLLLDIPLQTPHKLVDTGRAAKLIQRAEEMKSGLKDFKTFLTNDQTKV 471
Query 204 TE 205
TE
Sbjct 472 TE 473
> Hs19718755
Length=1765
Score = 108 bits (269), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 57/163 (34%), Positives = 100/163 (61%), Gaps = 17/163 (10%)
Query 11 QAPAQTAEDDEVANLEQEVTEGNWTRPEVEVHE-VKVETGEESEDTFWKCRSKLYRWAAG 69
Q+ D+E ++E +G + P V + + V+V +GEE+E + R+++YR+
Sbjct 1310 QSGTSVGTDEESVVTQEEERDGQYFEPVVPLPDLVEVSSGEENEQVVFSHRAEIYRYDKD 1369
Query 70 -GEWKERGLGEAKLLQHRETKKIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVS----- 123
G+WKERG+G+ K+LQ+ + K++R ++R+++ LK+ ANH ++TP++S
Sbjct 1370 VGQWKERGIGDIKILQNYDNKQVRIVMRRDQVLKLCANH---------RITPDMSLQNMK 1420
Query 124 -SEKIWVWTVMDFAEGELKNEQFALKFGQVEQAKEFKEKFEEA 165
+E++WVWT DFA+GE K E A++F + A FK+ F+EA
Sbjct 1421 GTERVWVWTACDFADGERKVEHLAVRFKLQDVADSFKKIFDEA 1463
Score = 103 bits (258), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 67/182 (36%), Positives = 104/182 (57%), Gaps = 16/182 (8%)
Query 37 PEVEVHE-VKVETGEESEDTFWKCRSKLYRWAA-GGEWKERGLGEAKLLQHRETKKIRFL 94
P V++ E V++ TGEE E + KL+R+ A +WKERGLG K+L++ K+R L
Sbjct 1039 PVVQMPEKVELVTGEEGEKVLYSQGVKLFRFDAEVSQWKERGLGNLKILKNEVNGKLRML 1098
Query 95 LRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAEGELKNEQFALKFGQVEQ 154
+R+E+ LK+ ANH++ T L P S++ W+W+ DF++G+ K E+ A KF E
Sbjct 1099 MRREQVLKVCANHWITTT---MNLKPLSGSDRAWMWSASDFSDGDAKLERLAAKFKTPEL 1155
Query 155 AKEFKEKFEEAAAI--------NSKLFGIGDSTKEGEKTEEKEA---EKSTESKDDETKE 203
A+EFK+KFEE + KL G + K ++ EE ++ + T +D+TK
Sbjct 1156 AEEFKQKFEECQRLLLDIPLQTPHKLVDTGRAAKLIQRAEEMKSGLKDFKTFLTNDQTKV 1215
Query 204 TE 205
TE
Sbjct 1216 TE 1217
> Hs13653066
Length=201
Score = 106 bits (265), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 60/133 (45%), Positives = 84/133 (63%), Gaps = 7/133 (5%)
Query 37 PEVEVHEVKVETGEESEDTFWKCRSKLYRWAAGG---EWKERGLGEAKLLQHRETKKIRF 93
P V + E +++T EE E+ +K R+K R+A+ EWKERG G+ KLL+H+E IR
Sbjct 29 PIVSLPEQEIKTLEEDEEELFKMRAKWLRFASENDLPEWKERGTGDVKLLKHKEKGAIRL 88
Query 94 LLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVW-TVMDFAEGELKNEQFALKFGQV 152
L++++KTLKI ANHY T +L PN S++ WVW T DFA+ K E A++F
Sbjct 89 LMQRDKTLKICANHY---TTPMMELKPNAGSDRAWVWNTHADFADECPKPELLAIRFLNA 145
Query 153 EQAKEFKEKFEEA 165
E A++FK KFEE
Sbjct 146 ENAQKFKTKFEEC 158
> At5g58590
Length=260
Score = 97.4 bits (241), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 62/167 (37%), Positives = 85/167 (50%), Gaps = 35/167 (20%)
Query 32 GNWTRPEVEVHEVKVETGEESEDT---FWKCRS----------KLYRWAA-GGEWKERGL 77
G P V + EV V TGEE ED + RS K+YR+ G +WKERG
Sbjct 24 GAQVAPIVRLEEVAVTTGEEDEDAVLDLYVTRSVNILVSLPLVKMYRFDKEGNQWKERGA 83
Query 78 GEAKLLQHRETKKIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAE 137
G KLL+H+ET K+R ++RQ KTLKI ANH + + + + +EK +W DF++
Sbjct 84 GTVKLLKHKETGKVRLVMRQSKTLKICANHLISSG---MSVQEHSGNEKSCLWHATDFSD 140
Query 138 GELKNEQFALKFGQVEQ------------------AKEFKEKFEEAA 166
GELK+E F ++F +E+ K F EKF E A
Sbjct 141 GELKDELFCIRFASIERKMVWKILEWLTDSVASTDCKTFMEKFTEIA 187
> CE17939
Length=860
Score = 86.7 bits (213), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 49/134 (36%), Positives = 82/134 (61%), Gaps = 5/134 (3%)
Query 36 RPEVEVHE-VKVETGEESEDTFWKCRSKLYRWAA-GGEWKERGLGEAKLLQHRETKKIRF 93
+P + + + V+V+TGEE E T + RSKLY +A EWKERG GE K+L +++ K R
Sbjct 256 KPVIPLPDLVEVKTGEEGEQTMFCNRSKLYIYANETKEWKERGTGELKVLYNKDKKSWRV 315
Query 94 LLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAEGELKNEQFALKFGQVE 153
++R+++ LK+ AN ++ + ++ S+EK + W DF+E + + + + +F V+
Sbjct 316 VMRRDQVLKVCANFPILGSMTIQQMK---SNEKAYTWFCEDFSEDQPAHVKLSARFANVD 372
Query 154 QAKEFKEKFEEAAA 167
A EFK FE+A A
Sbjct 373 IAGEFKTLFEKAVA 386
Score = 61.2 bits (147), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 42/134 (31%), Positives = 71/134 (52%), Gaps = 8/134 (5%)
Query 36 RPEVEVHE-VKVETGEESEDTFWKCRSKLYRWAAG-GEWKERGLGEAKLLQHRETKKIRF 93
+P + + + V+V+TGEE E+ + R KLY++ + E KERGLG+ KLL+ + K R
Sbjct 727 KPVIPLPDLVEVKTGEEDEEVMFSARCKLYKYYSDLKENKERGLGDIKLLKSND-NKYRI 785
Query 94 LLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAEGELKNEQ--FALKFGQ 151
++R+E+ K+ AN + + KL+P + + + DF+E + F KF
Sbjct 786 VMRREQVHKLCANFRI---EKSMKLSPKPNLPNVLTFMCQDFSEDASNADPAIFTAKFKD 842
Query 152 VEQAKEFKEKFEEA 165
A FK ++A
Sbjct 843 EATAGAFKTAVQDA 856
> Hs18579987
Length=147
Score = 69.3 bits (168), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 39/95 (41%), Positives = 53/95 (55%), Gaps = 3/95 (3%)
Query 71 EWKERGLGEAKLLQHRETKKIRFLLRQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVW 130
EWKERG G+ KLL+H+E IR L++++KTL I ANHY++ +L P
Sbjct 13 EWKERGTGDVKLLKHKEKGAIRLLMQRDKTLNICANHYIMR---MMELKPKQVVTVPGSG 69
Query 131 TVMDFAEGELKNEQFALKFGQVEQAKEFKEKFEEA 165
T M + K E A+ F E A++FK KFEE
Sbjct 70 TPMLTSPTIPKPELLAICFLNAENAQKFKTKFEEC 104
> At1g52380
Length=440
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 39/131 (29%), Positives = 63/131 (48%), Gaps = 10/131 (7%)
Query 30 TEGNWTRPEVEVHEVKVETGEESEDTFWKCRSKLYRWAAGGEWKERGLGEAKLLQHRETK 89
+EG+ P+ EV ETGEE+E + S ++ + GG WKERG GE K+
Sbjct 284 SEGSGFPPK---QEVSTETGEENEKVAFSADSIMFEYLDGG-WKERGKGELKVNVSSNDG 339
Query 90 KIRFLLRQEKTLKIVANHYVVATDVYCKL-TPNVSSEKIWVWTVMDFAEGELKNEQFALK 148
K R ++R + +++ N +Y ++ N+ + I V +EG+ FALK
Sbjct 340 KARLVMRAKGNYRLILN-----ASLYPEMKLANMDKKGITFACVNSVSEGKEGLSTFALK 394
Query 149 FGQVEQAKEFK 159
F +EF+
Sbjct 395 FKDPTIVEEFR 405
> YIL063c
Length=327
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 38/117 (32%), Positives = 64/117 (54%), Gaps = 8/117 (6%)
Query 46 VETGEESEDTFWKCRSKLYRWAAGGE-WKERGLGEAKLLQHR-ETKKIRFLLRQEKTLKI 103
V++GEESE+ ++ +KLY+ + E WKERG+G K+ + + + +K R ++R LK+
Sbjct 205 VKSGEESEECIYQVNAKLYQLSNIKEGWKERGVGIIKINKSKDDVEKTRIVMRSRGILKV 264
Query 104 VANHYVV-ATDVYCKLTPNVSSEK-IWVWTVMDFAEGELKNEQFALKFGQVEQAKEF 158
+ N +V V T ++ SEK I + V D + Q+A+K G+ E E
Sbjct 265 ILNIQLVKGFTVQKGFTGSLQSEKFIRLLAVDDNGD----PAQYAIKTGKKETTDEL 317
> At3g15970
Length=465
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 35/124 (28%), Positives = 57/124 (45%), Gaps = 6/124 (4%)
Query 42 HEVKVETGEESEDTFWKCRSKLYRWAAGGEWKERGLGEAKL-LQHRETKKIRFLLRQEKT 100
+V VETGEE+E + S ++ + GG WKERG GE K+ + E +K R ++R +
Sbjct 320 QDVSVETGEENEKAAFTADSVMFEYLEGG-WKERGKGELKVNISTTENRKARLVMRSKGN 378
Query 101 LKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAEGELKNEQFALKFGQVEQAKEFKE 160
++ N + K+ + I V ++ + ALKF +EF+
Sbjct 379 YRLTLNASLYPEMKLAKM----DKKGITFACVNSVSDAKDGLSTLALKFKDPTVVEEFRA 434
Query 161 KFEE 164
EE
Sbjct 435 VIEE 438
> SPCC18B5.07c
Length=565
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 36/59 (61%), Gaps = 1/59 (1%)
Query 49 GEESEDTFWKCRSKLYRW-AAGGEWKERGLGEAKLLQHRETKKIRFLLRQEKTLKIVAN 106
GEE+ED+ ++ R+K+YR+ A + + G+G K+ R+T R L R E + K++ N
Sbjct 448 GEENEDSVFETRAKIYRFDATSKSYSDIGIGPLKINVDRDTGSARILARVEGSGKLLLN 506
> Hs6466462
Length=499
Score = 37.4 bits (85), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 51/99 (51%), Gaps = 10/99 (10%)
Query 41 VHEVKVETGEESEDTFWKCRSKLYRWAAGGE-WKERGLGEAKLLQHRETK----KIRFLL 95
+ +V+V TGEE+E + + KL+ + + W ERG G +L T + R ++
Sbjct 318 LEKVEVITGEEAESNVLQMQCKLFVFDKTSQSWVERGRGLLRLNDMASTDDGTLQSRLVM 377
Query 96 RQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMD 134
R + +L+++ N T ++ ++ + +SEK T MD
Sbjct 378 RTQGSLRLILN-----TKLWAQMQIDKASEKSIRITAMD 411
> Hs6466466
Length=567
Score = 37.4 bits (85), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 51/99 (51%), Gaps = 10/99 (10%)
Query 41 VHEVKVETGEESEDTFWKCRSKLYRWAAGGE-WKERGLGEAKLLQHRETK----KIRFLL 95
+ +V+V TGEE+E + + KL+ + + W ERG G +L T + R ++
Sbjct 386 LEKVEVITGEEAESNVLQMQCKLFVFDKTSQSWVERGRGLLRLNDMASTDDGTLQSRLVM 445
Query 96 RQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMD 134
R + +L+++ N T ++ ++ + +SEK T MD
Sbjct 446 RTQGSLRLILN-----TKLWAQMQIDKASEKSIRITAMD 479
> Hs4506409
Length=562
Score = 37.4 bits (85), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 51/99 (51%), Gaps = 10/99 (10%)
Query 41 VHEVKVETGEESEDTFWKCRSKLYRWAAGGE-WKERGLGEAKLLQHRETK----KIRFLL 95
+ +V+V TGEE+E + + KL+ + + W ERG G +L T + R ++
Sbjct 381 LEKVEVITGEEAESNVLQMQCKLFVFDKTSQSWVERGRGLLRLNDMASTDDGTLQSRLVM 440
Query 96 RQEKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMD 134
R + +L+++ N T ++ ++ + +SEK T MD
Sbjct 441 RTQGSLRLILN-----TKLWAQMQIDKASEKSIRITAMD 474
> ECU02g0140
Length=219
Score = 32.0 bits (71), Expect = 0.98, Method: Compositional matrix adjust.
Identities = 29/126 (23%), Positives = 59/126 (46%), Gaps = 8/126 (6%)
Query 51 ESEDTFWKCRSKLYRWAAGGE-WKERGLGEAKLLQHRETKKIRFLLRQEKTLKIVANHYV 109
+S++ +K R KLY ++ + +ER G + H ++ + + +++ ++ NH++
Sbjct 88 QSDEILFKARCKLYYFSEETKALEERAEGTMIIEMHSKSNLAKITMFRDQIGRLGCNHFI 147
Query 110 VATDVYCKLTPNVSSEKIWVW-TVMDFAEGEL---KNEQFALKFGQVEQAKEFKEKFEEA 165
+ K P+ W+W + D E + K + F +KF E K F E+++
Sbjct 148 ---NPRFKAQPHGKVSNGWMWMSTEDTVETDALRKKIQLFVVKFYSEEDFKRFGEEYDLG 204
Query 166 AAINSK 171
A N K
Sbjct 205 RAHNEK 210
> 7293494
Length=872
Score = 30.8 bits (68), Expect = 2.3, Method: Composition-based stats.
Identities = 29/90 (32%), Positives = 43/90 (47%), Gaps = 10/90 (11%)
Query 134 DFAEGELKNEQFALKFGQVEQAKEFKEKFEEAAAINSKLFGIGDSTKEGEKTEEKEAE-- 191
DF + ++K + + V+Q KE ++ +EAA G T+E +K E E+E
Sbjct 502 DFMQ-DIKRLRAQINRNSVKQRKERAQETKEAATD-----GASTPTEEEDKDPEAESESK 555
Query 192 -KSTESKDDETKETEGAAAKTEAAAEVAKD 220
K+ E ET ETE AA TE + D
Sbjct 556 KKNQEPGSGET-ETESAADPTETPPSRSAD 584
> Hs14745035
Length=775
Score = 30.4 bits (67), Expect = 2.7, Method: Composition-based stats.
Identities = 28/112 (25%), Positives = 45/112 (40%), Gaps = 13/112 (11%)
Query 98 EKTLKIVANHYVVATDVYCKLTPNVSSEKIWVWTVMDFAEGELKNEQFALKFGQVEQAKE 157
+KT KI+ Y T CK P S K + + + L++ L VE+A
Sbjct 357 QKTSKIIQQEYEARTGRTCKPPPQSSRRKNFEFEPLSTTALILEDRPSNLPAKSVEEALR 416
Query 158 FKEKFEEAAAINSKLFGIGDSTKEGEKTEEKEAEKSTESKDDETKETEGAAA 209
+++++E A E +K E KEA K + K+ E A+
Sbjct 417 HRQEYDEMVA-------------EAKKREIKEAHKRKRIMKERFKQEENIAS 455
Lambda K H
0.305 0.122 0.339
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4419841864
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40