bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3133_orf2
Length=248
Score E
Sequences producing significant alignments: (Bits) Value
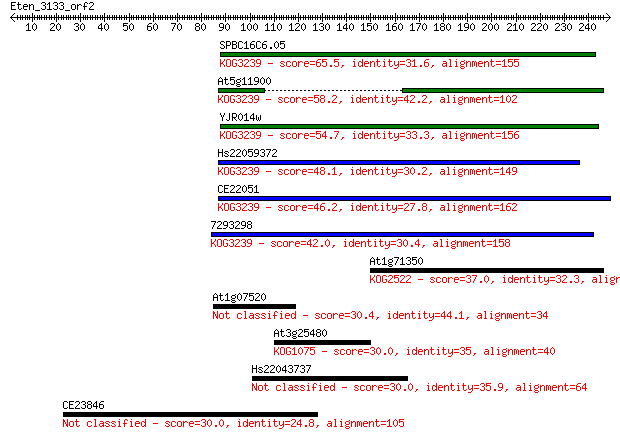
SPBC16C6.05 65.5 1e-10
At5g11900 58.2 2e-08
YJR014w 54.7 2e-07
Hs22059372 48.1 2e-05
CE22051 46.2 5e-05
7293298 42.0 0.001
At1g71350 37.0 0.038
At1g07520 30.4 3.6
At3g25480 30.0 4.4
Hs22043737 30.0 4.6
CE23846 30.0 4.6
> SPBC16C6.05
Length=190
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 49/175 (28%), Positives = 81/175 (46%), Gaps = 26/175 (14%)
Query 88 RKVEYCPTCGMPFEYCEFSGAACSKQEKDVKDPTQASVVQQNEHEQQLSDELQEKTAINE 147
+ V YC C +P EYCEF G K+ K+ + V + EQ LS +L+ ++
Sbjct 10 KSVLYCDVCTLPVEYCEFEGTL--KKCKEWLKSSHPDVYDKLYGEQDLSKDLENTLNVSG 67
Query 148 AKRGGA-------------------KKDGAKPKVVTVQKQAKRRGKCSTCVWGLEHFGVR 188
K A K+ +K + T+++ ++R T V GL+ FG+
Sbjct 68 TKDSNAEEQPAKLTKEEKRVEREEAKRMASKVLIKTIERTKRKR---VTTVQGLDAFGIE 124
Query 189 QEQAAKLASKHFACGASFQKGQPGQMPFVEIQGDVEETIAAFLNKNF-DIPGEKI 242
++AAK+ + FA GAS K + V +QGD+ I F+ + F ++P + I
Sbjct 125 TKKAAKMLANKFATGASVTKTADKKDEIV-VQGDLNYDIFDFILEKFKEVPEDNI 178
> At5g11900
Length=228
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/84 (36%), Positives = 47/84 (55%), Gaps = 2/84 (2%)
Query 163 VTVQKQAKRRGKCSTCVWGLEHFGVRQEQAAKLASKHFACGASFQKGQPGQMPFVEIQGD 222
V ++K + + KC T V GLE FG++ A+K K FA GAS KG P + +++QGD
Sbjct 137 VIIEKVVRNKRKCITIVKGLELFGIKLSDASKKLGKKFATGASVVKG-PTEKEQIDVQGD 195
Query 223 VEETIAAFLNKNF-DIPGEKIVFL 245
+ I F+ + D+P I F+
Sbjct 196 IIYDIVEFITDTWPDVPERSIFFI 219
Score = 33.9 bits (76), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 12/19 (63%), Positives = 13/19 (68%), Gaps = 0/19 (0%)
Query 87 PRKVEYCPTCGMPFEYCEF 105
P KV YC C +P EYCEF
Sbjct 7 PVKVLYCGVCSLPAEYCEF 25
> YJR014w
Length=198
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 52/180 (28%), Positives = 80/180 (44%), Gaps = 28/180 (15%)
Query 88 RKVEYCPTCGMPFEYCEFSGAA------CSKQEKDV-------KDPTQASVVQQNE---- 130
R+V YC C P EYCEFSG S+ D+ D TQ N+
Sbjct 3 REVIYCGICSYPPEYCEFSGKLKRCKVWLSENHADLYAKLYGTDDNTQEVEAVTNKLAES 62
Query 131 -----HEQQLSDELQ--EKTAINEAKRGGAKKDGAKPKVVTVQKQAKRRGKCSTCVWGLE 183
E++L +L +K N +R AKK +K V ++++A+ + K + GLE
Sbjct 63 SIGEAREEKLEKDLLKIQKKQENREQRELAKKLSSK---VIIKREARTKRKFIVAISGLE 119
Query 184 HFGVRQEQAAKLASKHFACGASFQKGQPGQMPFVEIQGDVEETIAAFLNKNFDIPGEKIV 243
F + ++ AK + FA G S K + V IQGDV + + +++ + G K V
Sbjct 120 VFDIDMKKLAKTFASRFATGCSVSKNAEKKEEVV-IQGDVMDEVETYIHSLLEEKGLKDV 178
> Hs22059372
Length=198
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 45/160 (28%), Positives = 75/160 (46%), Gaps = 16/160 (10%)
Query 87 PRKVEYCPTCGMPFEYCEFS-GAACSKQEKDVKDPTQ-ASVVQQNEHEQQLSDELQEKTA 144
P +V YC C +P EYCE+ A +Q + P + A + +N +Q+ + TA
Sbjct 28 PLRVLYCGVCSLPTEYCEYMPDVAKCRQWLEKNFPNEFAKLTVENSPKQEAGISEGQGTA 87
Query 145 -----INEAKRGG----AKKDGAKPKVVTVQKQAKRRGKCSTCVWGLEHFGVRQEQAAKL 195
+ KRGG +K P+ VT+ K + + K T V GL F + ++A +
Sbjct 88 GEEEEKKKQKRGGRGQIKQKKKTVPQKVTIAKIPRAKKKYVTRVCGLATFEIDLKEAQRF 147
Query 196 ASKHFACGASFQKGQPGQMPFVEIQGDVEETIAAFLNKNF 235
++ F+CGAS G+ + IQGD + I + + +
Sbjct 148 FAQKFSCGASVT----GEDEII-IQGDFTDDIIDVIQEKW 182
> CE22051
Length=192
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 45/174 (25%), Positives = 81/174 (46%), Gaps = 18/174 (10%)
Query 87 PRKVEYCPTCGMPFEYCEFSGAA-CSKQEKDVKDPTQASVVQQNEHEQQLSDELQEKTAI 145
P K+ YC C MP EYC++SG + P ++ ++ DE +++
Sbjct 23 PLKMVYCGQCSMPPEYCDYSGQTDVCRAWATQNAPELLEGLEISDEPAADGDEKKKQKRG 82
Query 146 NEAKRGG----------AKKDGAKPKVVTVQKQAKRRGKCSTCVWGLEHFGVRQEQAAKL 195
+ + G KK G P+ VT+Q++ + + K T + GL F + + A+KL
Sbjct 83 GKGSKTGAAAAQAAASGGKKKGGGPQKVTLQREPRGK-KSVTVIKGLATFDIDLKVASKL 141
Query 196 ASKHFACGASFQKGQPGQMPFVEIQGDVEETIAAFLNKNF-DIPGEKIVFLAEK 248
++ FACG+S + IQGDV++ + + + + + E+I L +K
Sbjct 142 FAQKFACGSSVTGADE-----IVIQGDVKDDLLDLIPEKWKQVTDEQIDDLGDK 190
> 7293298
Length=189
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 48/164 (29%), Positives = 71/164 (43%), Gaps = 21/164 (12%)
Query 84 MTGPRKVEYCPTCGMPFEYCEFSGAACS-KQEKDVKDPTQASVVQQNEHEQQLSDELQEK 142
+T P +++YC C MP EYCE+ K+ ++ P ++ E +K
Sbjct 23 VTYPIQMKYCGHCTMPIEYCEYYPEYEKCKEWLELHMPDDFERLKIEEEAAAADGTDDDK 82
Query 143 TAINEAKRGG-----AKKDGAKPKVVTVQKQAKRRGKCSTCVWGLEHFGVRQEQAAKLAS 197
KRGG KK PK + V + A+ + K T V GL F + + AAK
Sbjct 83 ---KRQKRGGKGLLRVKKKEDVPKRICVSRAARGKKKSVTVVTGLSTFDIDLKVAAKFFG 139
Query 198 KHFACGASFQKGQPGQMPFVEIQGDVEETIAAFLNKNFDIPGEK 241
FACG+S + IQGDV++ + FD+ EK
Sbjct 140 TKFACGSSVTGDDE-----IVIQGDVKDDL-------FDVIPEK 171
> At1g71350
Length=566
Score = 37.0 bits (84), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 31/99 (31%), Positives = 47/99 (47%), Gaps = 4/99 (4%)
Query 150 RGGAKKDGAKPKVVTVQKQAKRR--GKCSTCVWGLEHFGVRQEQAAKLASKHFACGASFQ 207
RGG + K V VQ +RR K T V G+E F + + K FAC S
Sbjct 464 RGGGEPVVRKGAVKPVQIMTERRQGNKKVTKVTGMETFLIDPDSFGSELQKKFACSTSVG 523
Query 208 KGQPGQMPF-VEIQGDVEETIAAFLNKNFDIPGEKIVFL 245
+ PG+ + V IQG V + +A ++ +++ +P I L
Sbjct 524 E-LPGKKGYEVLIQGGVIDNLARYMVEHYGVPKRYIEVL 561
> At1g07520
Length=695
Score = 30.4 bits (67), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 18/34 (52%), Gaps = 4/34 (11%)
Query 85 TGPRKVEYCPTCGMPFEYCEFSGAACSKQEKDVK 118
TG R EYC G+PFEY A SK + +K
Sbjct 478 TGRRLTEYCKRFGVPFEY----NAIASKNWETIK 507
> At3g25480
Length=1190
Score = 30.0 bits (66), Expect = 4.4, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 29/42 (69%), Gaps = 2/42 (4%)
Query 110 CSKQEKDVKDPTQASVVQQNE-HEQ-QLSDELQEKTAINEAK 149
C KQE+++K P+ +++++ E HE+ +L E++EK ++K
Sbjct 277 CQKQEENLKSPSPQAMIEETEAHEKWELLAEIEEKYLKQKSK 318
> Hs22043737
Length=380
Score = 30.0 bits (66), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 32/65 (49%), Gaps = 2/65 (3%)
Query 101 EYCEFSGAACSKQEKDVKDPTQASVVQQN-EHEQQLSDELQEKTAINEAKRGGAKKDGAK 159
E F S QE D + + ++QQ E +QQL + LQ + I EA G +KK G K
Sbjct 42 EVLHFREERLSLQENDSRLKHKLVLLQQQCEEKQQLFESLQSELQIYEALYGNSKK-GLK 100
Query 160 PKVVT 164
V+
Sbjct 101 DSAVS 105
> CE23846
Length=507
Score = 30.0 bits (66), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 26/109 (23%), Positives = 47/109 (43%), Gaps = 10/109 (9%)
Query 23 VGRIDGGDTNGQ----SMKRMEENELTSNQDDLGAADASSNPPALGQHSDAETSISCWEH 78
+ + D + N Q + +R+EE S DD+GA ++ A + ++T +
Sbjct 306 ISKYDRSNGNAQGFLVTAQRLEEE---SKSDDIGAGCVTAVELACKIRNQSKTIMDT--I 360
Query 79 DRQPPMTGPRKVEYCPTCGMPFEYCEFSGAACSKQEKDVKDPTQASVVQ 127
R P + GPR ++ CG+ +E E C + +PT V+
Sbjct 361 TRHPSL-GPRMMQLAERCGVGYENEEIIHQICDRFSLHPSEPTWLDYVE 408
Lambda K H
0.312 0.128 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5045397172
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40