bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3132_orf2
Length=187
Score E
Sequences producing significant alignments: (Bits) Value
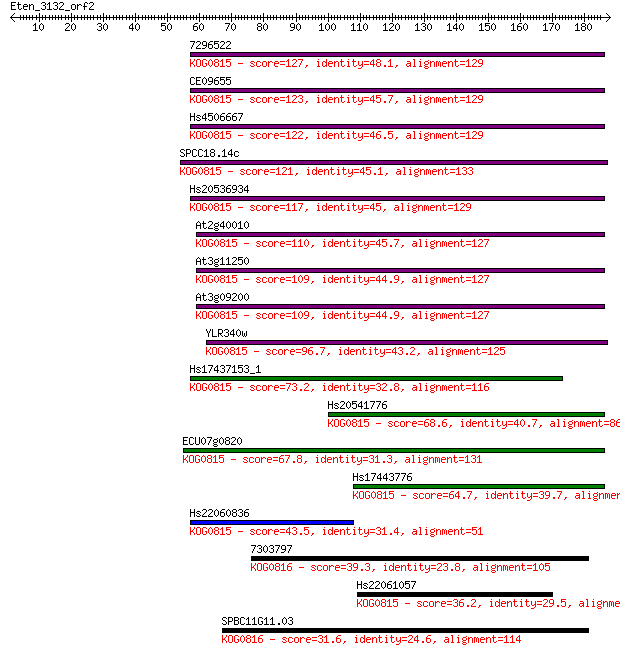
7296522 127 9e-30
CE09655 123 2e-28
Hs4506667 122 6e-28
SPCC18.14c 121 1e-27
Hs20536934 117 1e-26
At2g40010 110 2e-24
At3g11250 109 3e-24
At3g09200 109 3e-24
YLR340w 96.7 3e-20
Hs17437153_1 73.2 3e-13
Hs20541776 68.6 6e-12
ECU07g0820 67.8 1e-11
Hs17443776 64.7 1e-10
Hs22060836 43.5 3e-04
7303797 39.3 0.005
Hs22061057 36.2 0.035
SPBC11G11.03 31.6 0.89
> 7296522
Length=317
Score = 127 bits (320), Expect = 9e-30, Method: Compositional matrix adjust.
Identities = 62/129 (48%), Positives = 87/129 (67%), Gaps = 0/129 (0%)
Query 57 KERAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRT 116
+ +AA + +YF ++++L E P+ +V AD+VGSKQ+ IR +LRG A VLMGKNT +R
Sbjct 4 ENKAAWKAQYFIKVVELFDEFPKCFIVGADNVGSKQMQNIRTSLRGLAVVLMGKNTMMRK 63
Query 117 ALRQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFI 176
A+R L+ PQL+ LLP ++ NVGF+F + D A VR + + KV APA+ G AP V I
Sbjct 64 AIRGHLENNPQLEKLLPHIKGNVGFVFTKGDLAEVRDKLLESKVRAPARPGAIAPLHVII 123
Query 177 PAGPTGMDP 185
PA TG+ P
Sbjct 124 PAQNTGLGP 132
> CE09655
Length=312
Score = 123 bits (309), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 59/129 (45%), Positives = 85/129 (65%), Gaps = 0/129 (0%)
Query 57 KERAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRT 116
++R+ + YF +L++L E+P+ L+V D+VGSKQ+ IR A+RG A +LMGKNT IR
Sbjct 4 EDRSTWKANYFTKLVELFEEYPKCLLVGVDNVGSKQMQEIRQAMRGHAEILMGKNTMIRK 63
Query 117 ALRQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFI 176
ALR L + P L+ LLP + NVGF+F + D +R+ + +++ APAK G AP DV +
Sbjct 64 ALRGHLGKNPSLEKLLPHIVENVGFVFTKEDLGEIRSKLLENRKGAPAKAGAIAPCDVKL 123
Query 177 PAGPTGMDP 185
P TGM P
Sbjct 124 PPQNTGMGP 132
> Hs4506667
Length=317
Score = 122 bits (305), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 60/129 (46%), Positives = 90/129 (69%), Gaps = 0/129 (0%)
Query 57 KERAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRT 116
++RA + YF +++QLL ++P+ +V AD+VGSKQ+ IR++LRG+A VLMGKNT +R
Sbjct 4 EDRATWKSNYFLKIIQLLDDYPKCFIVGADNVGSKQMQQIRMSLRGKAVVLMGKNTMMRK 63
Query 117 ALRQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFI 176
A+R L+ P L+ LLP +R NVGF+F + D +R ++ +KVPA A+ G AP +V +
Sbjct 64 AIRGHLENNPALEKLLPHIRGNVGFVFTKEDLTEIRDMLLANKVPAAARAGAIAPCEVTV 123
Query 177 PAGPTGMDP 185
PA TG+ P
Sbjct 124 PAQNTGLGP 132
> SPCC18.14c
Length=312
Score = 121 bits (303), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 60/133 (45%), Positives = 85/133 (63%), Gaps = 3/133 (2%)
Query 54 AMGKERAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTK 113
A+ KE A +YF +L L ++ + VV+ D+V S+Q+ +R LRG A ++MGKNT
Sbjct 2 AISKESKA---QYFEKLRSLFEKYNSLFVVNIDNVSSQQMHTVRKQLRGTAELIMGKNTM 58
Query 114 IRTALRQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTD 173
IR A+R + P+L+ LLP+VR NVGF+F AD VR + + + APA+ AP D
Sbjct 59 IRRAMRGIINDMPELERLLPVVRGNVGFVFTNADLKEVRETIIANVIAAPARPNAIAPLD 118
Query 174 VFIPAGPTGMDPG 186
VF+PAG TGM+PG
Sbjct 119 VFVPAGNTGMEPG 131
> Hs20536934
Length=317
Score = 117 bits (294), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 58/129 (44%), Positives = 88/129 (68%), Gaps = 0/129 (0%)
Query 57 KERAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRT 116
++RA + YF +++QLL ++P+ +V AD+VGSKQ+ IR++LRG+ VLMGKNT +R
Sbjct 4 EDRATWKSNYFLKIIQLLDDYPKCFIVGADNVGSKQMQQIRMSLRGKVVVLMGKNTMMRK 63
Query 117 ALRQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFI 176
A+R L+ P L+ LLP + NVGF+F + D +R ++ +KVPA A+ G AP +V +
Sbjct 64 AIRGHLENNPALEKLLPHIWGNVGFVFTKEDLTEIRDMLLANKVPAAARAGAIAPCEVTV 123
Query 177 PAGPTGMDP 185
PA TG+ P
Sbjct 124 PAQNTGLGP 132
> At2g40010
Length=317
Score = 110 bits (275), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 58/129 (44%), Positives = 82/129 (63%), Gaps = 2/129 (1%)
Query 59 RAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRTAL 118
+A K+ Y +L QLL E+ ++LVV+AD+VGS QL IR LRG + VLMGKNT ++ ++
Sbjct 7 KAEKKIVYDSKLCQLLNEYSQILVVAADNVGSTQLQNIRKGLRGDSVVLMGKNTMMKRSV 66
Query 119 RQQLQQQPQ--LQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFI 176
R + +LLPL++ NVG IF + D V V ++KV APA+ G+ AP DV +
Sbjct 67 RIHADKTGNQAFLSLLPLLQGNVGLIFTKGDLKEVSEEVAKYKVGAPARVGLVAPIDVVV 126
Query 177 PAGPTGMDP 185
G TG+DP
Sbjct 127 QPGNTGLDP 135
> At3g11250
Length=323
Score = 109 bits (273), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 57/129 (44%), Positives = 82/129 (63%), Gaps = 2/129 (1%)
Query 59 RAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRTAL 118
+A K+ Y +L QL+ E+ ++LVV+AD+VGS QL IR LRG + VLMGKNT ++ ++
Sbjct 6 KAEKKIAYDTKLCQLIDEYTQILVVAADNVGSTQLQNIRKGLRGDSVVLMGKNTMMKRSV 65
Query 119 RQQLQQ--QPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFI 176
R + + LLPL++ NVG IF + D V V ++KV APA+ G+ AP DV +
Sbjct 66 RIHSENSGNTAILNLLPLLQGNVGLIFTKGDLKEVSEEVAKYKVGAPARVGLVAPIDVVV 125
Query 177 PAGPTGMDP 185
G TG+DP
Sbjct 126 QPGNTGLDP 134
> At3g09200
Length=320
Score = 109 bits (272), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 57/129 (44%), Positives = 82/129 (63%), Gaps = 2/129 (1%)
Query 59 RAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRTAL 118
+A K+ Y +L QL+ E+ ++LVV+AD+VGS QL IR LRG + VLMGKNT ++ ++
Sbjct 6 KAEKKIAYDTKLCQLIDEYTQILVVAADNVGSTQLQNIRKGLRGDSVVLMGKNTMMKRSV 65
Query 119 RQQLQQ--QPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFI 176
R + + LLPL++ NVG IF + D V V ++KV APA+ G+ AP DV +
Sbjct 66 RIHSENTGNTAILNLLPLLQGNVGLIFTKGDLKEVSEEVAKYKVGAPARVGLVAPIDVVV 125
Query 177 PAGPTGMDP 185
G TG+DP
Sbjct 126 QPGNTGLDP 134
> YLR340w
Length=312
Score = 96.7 bits (239), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 54/125 (43%), Positives = 79/125 (63%), Gaps = 0/125 (0%)
Query 62 KRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRTALRQQ 121
K+ EYF +L + L E+ + VV D+V S+Q+ +R LRG+A VLMGKNT +R A+R
Sbjct 7 KKAEYFAKLREYLEEYKSLFVVGVDNVSSQQMHEVRKELRGRAVVLMGKNTMVRRAIRGF 66
Query 122 LQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFIPAGPT 181
L P + LLP V+ NVGF+F ++ V+ ++V APA+ G AP D+++ A T
Sbjct 67 LSDLPDFEKLLPFVKGNVGFVFTNEPLTEIKNVIVSNRVAAPARAGAVAPEDIWVRAVNT 126
Query 182 GMDPG 186
GM+PG
Sbjct 127 GMEPG 131
> Hs17437153_1
Length=334
Score = 73.2 bits (178), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 38/116 (32%), Positives = 63/116 (54%), Gaps = 22/116 (18%)
Query 57 KERAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRT 116
++R + YF +++QLL ++P+ ++ AD++GSKQ+ IR++L +A VL+GKNT +
Sbjct 132 EDRVTWKPNYFLKIIQLLDDYPKCFIMGADNMGSKQMQQIRISLCRKAMVLLGKNTTMG- 190
Query 117 ALRQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPT 172
NVGF+F + D +R ++ +KVPA A G AP
Sbjct 191 ---------------------NVGFVFIKEDLTEIRDLLLANKVPAVAHAGSIAPV 225
> Hs20541776
Length=234
Score = 68.6 bits (166), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 35/86 (40%), Positives = 52/86 (60%), Gaps = 0/86 (0%)
Query 100 LRGQATVLMGKNTKIRTALRQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHK 159
+ G+A V MG+N +R A + QL+ P L+ LLP ++ NVGF F + D VR ++ +K
Sbjct 27 MEGKAVVPMGRNPMMRKATQGQLENSPALEKLLPPLQGNVGFAFTKEDLTEVRDLLLANK 86
Query 160 VPAPAKQGVTAPTDVFIPAGPTGMDP 185
VPA + G P +V +PA TG+ P
Sbjct 87 VPAATRAGAIGPCEVTVPAQNTGLGP 112
> ECU07g0820
Length=290
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 41/132 (31%), Positives = 71/132 (53%), Gaps = 3/132 (2%)
Query 55 MGKERAAKRREY-FPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTK 113
M ++ A +R+E + R +L + R +V ++V S QL I+ G A +LMGKN+
Sbjct 25 MTRKDAKERKELTYERARKLFETYSRFALVKIENVVSTQLKDIKRQWGGNAELLMGKNSA 84
Query 114 IRTALRQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTD 173
IR A+ +P+L + LV+ +V F+F + + ++ + ++ A AK G A D
Sbjct 85 IRRAIAD--LGKPELSGVYDLVKGDVCFVFFKGNARDIKKAIDENVREACAKVGNVAQRD 142
Query 174 VFIPAGPTGMDP 185
V++ + TGM P
Sbjct 143 VWVESCITGMTP 154
> Hs17443776
Length=158
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 31/78 (39%), Positives = 45/78 (57%), Gaps = 0/78 (0%)
Query 108 MGKNTKIRTALRQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQG 167
MGKNT + A++ L+ P L+ LLP ++ NVGF F + D R ++ KVPA G
Sbjct 4 MGKNTMMCKAIQGHLENNPALEKLLPHIQGNVGFAFTKEDLTKFRDMLLNSKVPAATHAG 63
Query 168 VTAPTDVFIPAGPTGMDP 185
AP +V +P TG++P
Sbjct 64 AIAPCEVTVPGQNTGLEP 81
> Hs22060836
Length=137
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 36/51 (70%), Gaps = 0/51 (0%)
Query 57 KERAAKRREYFPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVL 107
++RA + YF ++++ L ++P+ +V ++VGS+Q+ I+++L+G+A L
Sbjct 4 EDRATWKSNYFLKVIEHLDDYPKCFIVGVNNVGSEQMKQIQMSLQGKAEDL 54
> 7303797
Length=256
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 25/107 (23%), Positives = 48/107 (44%), Gaps = 2/107 (1%)
Query 76 EHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRTAL-RQQLQQ-QPQLQALLP 133
++P + V ++ + L +R L+ + + GKN ++ L R + ++ +P+L L
Sbjct 36 KYPNIFVFQVQNMRNSLLKDLRQELKKNSRFIFGKNRVMQIGLGRTKSEEVEPELHKLSK 95
Query 134 LVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFIPAGP 180
+ VG +F V + + A+ G A V +PAGP
Sbjct 96 RLTGQVGLLFTDKSKEEVLEWAENYWAVEYARSGFVATETVTLPAGP 142
> Hs22061057
Length=184
Score = 36.2 bits (82), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 32/61 (52%), Gaps = 0/61 (0%)
Query 109 GKNTKIRTALRQQLQQQPQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGV 168
G+ T + A++ L+ L+ LLP ++ NVGF+F + D +R ++ K + G
Sbjct 30 GQETMMLKAIQGHLENSRALEKLLPRIQGNVGFVFTKEDLTEIRDMLVASKASSLKPLGT 89
Query 169 T 169
T
Sbjct 90 T 90
> SPBC11G11.03
Length=241
Score = 31.6 bits (70), Expect = 0.89, Method: Compositional matrix adjust.
Identities = 28/116 (24%), Positives = 47/116 (40%), Gaps = 3/116 (2%)
Query 67 FPRLLQLLLEHPRVLVVSADHVGSKQLAGIRVALRGQATVLMGKNTKIRTALRQQLQQQ- 125
F + Q L + + ++ + L IR +G + + MGK + AL +++
Sbjct 27 FSGVQQSLDSFDYMWIFDVTNMRNTYLKRIRDDWKG-SRIFMGKTKVMAKALGHTPEEEH 85
Query 126 -PQLQALLPLVRLNVGFIFCRADPAAVRAVVQQHKVPAPAKQGVTAPTDVFIPAGP 180
+ L L+ VG +F + P V + A+ G AP IPAGP
Sbjct 86 AENVSKLTKLLHGAVGLLFTNSKPDEVIGYFESFVQNDFARAGAVAPFTHVIPAGP 141
Lambda K H
0.321 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3058524910
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40