bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3129_orf3
Length=301
Score E
Sequences producing significant alignments: (Bits) Value
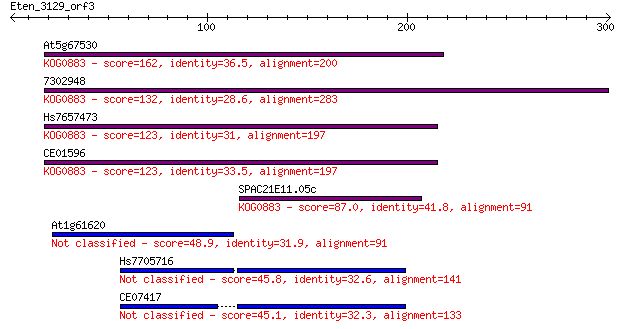
At5g67530 162 1e-39
7302948 132 8e-31
Hs7657473 123 4e-28
CE01596 123 5e-28
SPAC21E11.05c 87.0 5e-17
At1g61620 48.9 1e-05
Hs7705716 45.8 1e-04
CE07417 45.1 2e-04
> At5g67530
Length=595
Score = 162 bits (409), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 73/200 (36%), Positives = 117/200 (58%), Gaps = 22/200 (11%)
Query 18 MPKHKHHRDRLYLLQSEYSSEWGGFKKAKLNLPYKVLPFYCCCLCFCPAKDPVCSSSGAV 77
M K +H +DR+++ ++E+++EWGG K + P+K LP+YCC L F P +DPVC+ G+V
Sbjct 1 MGKKQHSKDRMFITKTEWATEWGGAKSKENRTPFKSLPYYCCALTFLPFEDPVCTIDGSV 60
Query 78 FDRSCILSFMQQQQKLQQKQQQQQQQQQQQQQELECQCPVTGQRLRFADLFALKFFKNAK 137
F+ + I+ ++++ K PVTG L+ DL L F KN++
Sbjct 61 FEITTIVPYIRKFGK----------------------HPVTGAPLKGEDLIPLIFHKNSE 98
Query 138 GEICCPVSFKLLSENSHVAAVRSSGNVFAADTLLNLCVRTNSWNDLITGEPFTSSDIVHI 197
GE CPV K+ +E +H+ AV+++GNVF + + L ++T +W +L+T EPFT +D++ I
Sbjct 99 GEYHCPVLNKVFTEFTHIVAVKTTGNVFCYEAIKELNIKTKNWKELLTEEPFTRADLITI 158
Query 198 QNPTDAKRRCIEHFHHVLKG 217
QNP + F HV G
Sbjct 159 QNPNAVDGKVTVEFDHVKNG 178
> 7302948
Length=517
Score = 132 bits (332), Expect = 8e-31, Method: Compositional matrix adjust.
Identities = 81/286 (28%), Positives = 133/286 (46%), Gaps = 35/286 (12%)
Query 18 MPKHKHHRDRLYLLQSEYSSEWGGFKKAKL---NLPYKVLPFYCCCLCFCPAKDPVCSSS 74
M K +H +D++YL +E+S +GG K L ++ +K LPF CC+ P + P C
Sbjct 1 MGKRQHQKDKMYLTYTEWSELYGGKKVESLENDHVKFKRLPFEHCCITMAPYEMPYCDLQ 60
Query 75 GAVFDRSCILSFMQQQQKLQQKQQQQQQQQQQQQQELECQCPVTGQRLRFADLFALKFFK 134
G VF+ IL F++ + P+TGQ++ L L F +
Sbjct 61 GNVFEYEAILKFLKTFK----------------------VNPITGQKMDSKSLVKLNFHR 98
Query 135 NAKGEICCPVSFKLLSENSHVAAVRSSGNVFAADTLLNLCVRTNSWNDLITGEPFTSSDI 194
NA E CP FK S+NSH+ AV ++GNV+ + + L ++T +W DL+ PF DI
Sbjct 99 NANDEYHCPALFKPFSKNSHIVAVATTGNVYCWEAIDQLNIKTKNWKDLVDDTPFQRKDI 158
Query 195 VHIQNPTDAKRRCIEHFHHVLKGGGSAQLVRGVASEEGSKVPSIKKNESMLKVLAEAAKT 254
+ IQ+P ++ I F+H+ K +R + EE + +KN + ++ +T
Sbjct 159 ITIQDPQKLEKYDISTFYHIKKN------LRVLTEEEQQE----RKNPASGRIKTMNLET 208
Query 255 RAAVEAAHNRIVPNMNAPAAAAAAAAAPAAAAPAAAAPAAAAPAAA 300
+ +E P + + A AA + A AA+ + A
Sbjct 209 KETLEQLQQDYQPAEEEASTSKRTADKFNAAHYSTGAVAASFTSTA 254
> Hs7657473
Length=520
Score = 123 bits (309), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 61/197 (30%), Positives = 105/197 (53%), Gaps = 22/197 (11%)
Query 18 MPKHKHHRDRLYLLQSEYSSEWGGFKKAKLNLPYKVLPFYCCCLCFCPAKDPVCSSSGAV 77
M K +H +D++Y+ +EY+ +GG K ++ LPF C L P PVC+ G V
Sbjct 1 MGKRQHQKDKMYITCAEYTHFYGGKKPDLPQTNFRRLPFDHCSLSLQPFVYPVCTPDGIV 60
Query 78 FDRSCILSFMQQQQKLQQKQQQQQQQQQQQQQELECQCPVTGQRLRFADLFALKFFKNAK 137
FD I+ ++++ P G++L L L F KN++
Sbjct 61 FDLLNIVPWLKKY----------------------GTNPSNGEKLDGRSLIKLNFSKNSE 98
Query 138 GEICCPVSFKLLSENSHVAAVRSSGNVFAADTLLNLCVRTNSWNDLITGEPFTSSDIVHI 197
G+ CPV F + + N+H+ AVR++GNV+A + + L ++ ++ DL+T EPF+ DI+ +
Sbjct 99 GKYHCPVLFTVFTNNTHIVAVRTTGNVYAYEAVEQLNIKAKNFRDLLTDEPFSRQDIITL 158
Query 198 QNPTDAKRRCIEHFHHV 214
Q+PT+ + + +F+HV
Sbjct 159 QDPTNLDKFNVSNFYHV 175
> CE01596
Length=523
Score = 123 bits (309), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 66/200 (33%), Positives = 104/200 (52%), Gaps = 25/200 (12%)
Query 18 MPKHKHHRDRLYLLQSEYSSEWGGFKKAKLNLP---YKVLPFYCCCLCFCPAKDPVCSSS 74
M K +H +D+LYL SE+ S G L +K LP C L P +DPVC+ S
Sbjct 1 MGKKQHQKDKLYLTTSEWKSIGGHKDDTGTRLQRAQFKRLPINHCSLSLLPFEDPVCARS 60
Query 75 GAVFDRSCILSFMQQQQKLQQKQQQQQQQQQQQQQELECQCPVTGQRLRFADLFALKFFK 134
G +FD + I+ ++++ K P TG+ L DL LKF K
Sbjct 61 GEIFDLTAIVPYLKKHGK----------------------NPCTGKPLVAKDLIHLKFDK 98
Query 135 NAKGEICCPVSFKLLSENSHVAAVRSSGNVFAADTLLNLCVRTNSWNDLITGEPFTSSDI 194
G+ CPV+F+ +++SH+ A+ +SGNV++ + + L ++ N DL+T PFT +DI
Sbjct 99 GEDGKFRCPVTFRTFTDHSHILAIATSGNVYSHEAVQELNLKRNHLKDLLTDVPFTRADI 158
Query 195 VHIQNPTDAKRRCIEHFHHV 214
+ +Q+P ++ +E F HV
Sbjct 159 IDLQDPNHLEKFNMEQFLHV 178
> SPAC21E11.05c
Length=471
Score = 87.0 bits (214), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 38/91 (41%), Positives = 56/91 (61%), Gaps = 0/91 (0%)
Query 116 PVTGQRLRFADLFALKFFKNAKGEICCPVSFKLLSENSHVAAVRSSGNVFAADTLLNLCV 175
P+ GQ+ +DL LKF KN+ E C PV+ K + SH+ A+RS+GN F+ DT+ L +
Sbjct 39 PINGQKASMSDLIKLKFAKNSAEEYCDPVTMKSFTRFSHIVAIRSTGNCFSWDTIERLNI 98
Query 176 RTNSWNDLITGEPFTSSDIVHIQNPTDAKRR 206
+ W DL+ E FT DI+ IQ+P + + R
Sbjct 99 KPKHWRDLVNEEQFTRDDIITIQDPHNVENR 129
> At1g61620
Length=310
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/92 (31%), Positives = 45/92 (48%), Gaps = 2/92 (2%)
Query 22 KHHRDRLYLLQSEYSSEWGGFKKAKLNLPYKVLPFYCCCLCFCPAKDPVCSSSGAVFDRS 81
K++ D Y E G ++ +L + PF C LC P DP+C G VF R
Sbjct 7 KNNNDLAYFTYDEKKKLGYGTQRERLGRD-SIKPFDACSLCLKPFIDPMCCHKGHVFCRE 65
Query 82 CILS-FMQQQQKLQQKQQQQQQQQQQQQQELE 112
CIL F+ Q++ +Q++ Q++Q + E E
Sbjct 66 CILECFLAQKKDIQRRLAAHSSQKKQDKDEEE 97
> Hs7705716
Length=301
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 37/61 (60%), Gaps = 4/61 (6%)
Query 56 FYCCCLCFCPAKDPVCSSSGAVFDRSCILSFMQQQQKLQQKQ----QQQQQQQQQQQQEL 111
F CCCL P DPV + G +++R IL ++ Q+K +Q ++Q+ ++++Q+EL
Sbjct 43 FDCCCLSLQPCHDPVVTPDGYLYEREAILEYILHQKKEIARQMKAYEKQRGTRREEQKEL 102
Query 112 E 112
+
Sbjct 103 Q 103
Score = 35.8 bits (81), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 26/95 (27%), Positives = 42/95 (44%), Gaps = 13/95 (13%)
Query 115 CPVTGQRLRFADLFALKF-----------FKNAKGEICCPVSFKLLSENSHVAAVRSSGN 163
CP++G+ LR +DL + F C V+ LS + A +R SG
Sbjct 185 CPMSGKPLRMSDLTPVHFTPLDSSVDRVGLITRSERYVCAVTRDSLSNATPCAVLRPSGA 244
Query 164 VFAADTLLNLCVRTNSWNDLITGEPFTSSDIVHIQ 198
V + + L +R + D +TG+ T DI+ +Q
Sbjct 245 VVTLECVEKL-IRKD-MVDPVTGDKLTDRDIIVLQ 277
> CE07417
Length=310
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 30/49 (61%), Gaps = 0/49 (0%)
Query 56 FYCCCLCFCPAKDPVCSSSGAVFDRSCILSFMQQQQKLQQKQQQQQQQQ 104
F+CC L P ++PV S +G +FDR IL + Q+K K+ ++ ++Q
Sbjct 43 FHCCSLTLQPCRNPVISPTGYIFDREAILENILAQKKAYAKKLKEYEKQ 91
Score = 43.5 bits (101), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 25/90 (27%), Positives = 42/90 (46%), Gaps = 8/90 (8%)
Query 115 CPVTGQRLRFADLFALKFF------KNAKGEICCPVSFKLLSENSHVAAVRSSGNVFAAD 168
CPV+G+ ++ +L +KF A + CPV+ L+ + A ++ S +V D
Sbjct 199 CPVSGKPIKLKELLEVKFTPMPGTETAAHRKFLCPVTRDELTNTTRCAYLKKSKSVVKYD 258
Query 169 TLLNLCVRTNSWNDLITGEPFTSSDIVHIQ 198
+ L D I GEP + DI+ +Q
Sbjct 259 VVEKLI--KGDGIDPINGEPMSEDDIIELQ 286
Lambda K H
0.319 0.129 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6813889318
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40