bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3113_orf3
Length=221
Score E
Sequences producing significant alignments: (Bits) Value
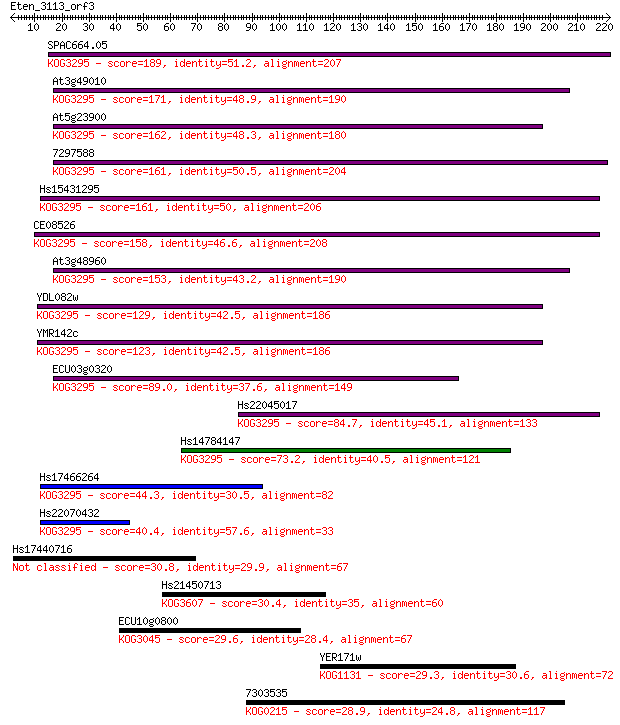
SPAC664.05 189 4e-48
At3g49010 171 2e-42
At5g23900 162 3e-40
7297588 161 8e-40
Hs15431295 161 1e-39
CE08526 158 1e-38
At3g48960 153 3e-37
YDL082w 129 7e-30
YMR142c 123 2e-28
ECU03g0320 89.0 6e-18
Hs22045017 84.7 1e-16
Hs14784147 73.2 4e-13
Hs17466264 44.3 2e-04
Hs22070432 40.4 0.003
Hs17440716 30.8 2.5
Hs21450713 30.4 2.6
ECU10g0800 29.6 4.9
YER171w 29.3 6.3
7303535 28.9 9.1
> SPAC664.05
Length=208
Score = 189 bits (480), Expect = 4e-48, Method: Compositional matrix adjust.
Identities = 106/210 (50%), Positives = 137/210 (65%), Gaps = 11/210 (5%)
Query 15 AQLPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPLGLLRPAVRCPTQKYN 74
QLPN H K+WQR+V+TWF+QP RK RRR RQ KAAK+ RP+ +RPAV+ PT +YN
Sbjct 7 GQLPNAHFHKDWQRYVKTWFNQPGRKLRRRQARQTKAAKIAPRPVEAIRPAVKPPTIRYN 66
Query 75 IKLREGRGFTLEELKAVGISPRVALSIGISVDHRRRNRCQESLNTNVNRLKLYLSKLVLF 134
+K+R GRGFTLEELKA G+S RVA +IGI VDHRRRNR +ESL NV R+K+YL+ L++F
Sbjct 67 MKVRAGRGFTLEELKAAGVSRRVASTIGIPVDHRRRNRSEESLQRNVERIKVYLAHLIVF 126
Query 135 QRGSKAKKGLAGVPADTPKSQLQNVKQAKIADSLPIERPRKKIRARVITEEERKFRAYAT 194
R AG P + + +Q +A LPI + + A+ ITEE + F A++T
Sbjct 127 PRK-------AGQPKKGDATDVSGAEQTDVAAVLPITQEAVE-EAKPITEEAKNFNAFST 178
Query 195 LRKQLRDAKNVGKHA---KKLAEEAEAANK 221
L + A+ G A KK AEEAEA K
Sbjct 179 LSNERAYARYAGARAAFQKKRAEEAEAKKK 208
> At3g49010
Length=206
Score = 171 bits (432), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 93/191 (48%), Positives = 126/191 (65%), Gaps = 7/191 (3%)
Query 17 LPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPL-GLLRPAVRCPTQKYNI 75
+PN H +K WQ +V+TWF+QPARK RRR+ RQKKA K+ RP G LRP V T KYN+
Sbjct 7 IPNGHFKKHWQNYVKTWFNQPARKTRRRIARQKKAVKIFPRPTSGPLRPVVHGQTLKYNM 66
Query 76 KLREGRGFTLEELKAVGISPRVALSIGISVDHRRRNRCQESLNTNVNRLKLYLSKLVLFQ 135
K+R G+GFTLEELKA GI ++A +IGI+VDHRR+NR E L TNV RLK Y +KLV+F
Sbjct 67 KVRTGKGFTLEELKAAGIPKKLAPTIGIAVDHRRKNRSLEGLQTNVQRLKTYKTKLVIFP 126
Query 136 RGSKAKKGLAGVPADTPKSQLQNVKQAKIADSLPIERPRKKIRARVITEEERKFRAYATL 195
R +A+K AG D+ +L N Q + D LPI R + + +T E + F+A+ +
Sbjct 127 R--RARKVKAG---DSTPEELANATQVQ-GDYLPIVREKPTMELVKLTSEMKSFKAFDKI 180
Query 196 RKQLRDAKNVG 206
R + + ++ G
Sbjct 181 RLERTNKRHAG 191
> At5g23900
Length=206
Score = 162 bits (411), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 87/181 (48%), Positives = 116/181 (64%), Gaps = 7/181 (3%)
Query 17 LPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPL-GLLRPAVRCPTQKYNI 75
+P+ H RK WQ +V+TWF+QPARK RRR+ RQKKA K+ RP G LRP V T KYN+
Sbjct 7 IPSSHFRKHWQNYVKTWFNQPARKTRRRVARQKKAVKIFPRPTSGPLRPVVHGQTLKYNM 66
Query 76 KLREGRGFTLEELKAVGISPRVALSIGISVDHRRRNRCQESLNTNVNRLKLYLSKLVLFQ 135
K+R G+GFTLEELK GI ++A +IGISVDHRR+NR E L +NV RLK Y +KLV+F
Sbjct 67 KVRAGKGFTLEELKVAGIPKKLAPTIGISVDHRRKNRSLEGLQSNVQRLKTYKAKLVVFP 126
Query 136 RGSKAKKGLAGVPADTPKSQLQNVKQAKIADSLPIERPRKKIRARVITEEERKFRAYATL 195
R S+ K D+ +L N Q + D +PI + + +T + + F+AY +
Sbjct 127 RRSRQVKA-----GDSTPEELANATQVQ-GDYMPIASVKAAMELVKLTADLKAFKAYDKI 180
Query 196 R 196
R
Sbjct 181 R 181
> 7297588
Length=218
Score = 161 bits (408), Expect = 8e-40, Method: Compositional matrix adjust.
Identities = 103/209 (49%), Positives = 129/209 (61%), Gaps = 14/209 (6%)
Query 17 LPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPL-GLLRPAVRCPTQKYNI 75
+PN H K WQR V+TWF+QPARK RR R KKA + RP G LRP VRCPT +Y+
Sbjct 8 IPNQHYHKWWQRHVKTWFNQPARKVRRHANRVKKAKAVFPRPASGALRPVVRCPTIRYHT 67
Query 76 KLREGRGFTLEELKAVGISPRVALSIGISVDHRRRNRCQESLNTNVNRLKLYLSKLVLFQ 135
KLR GRGFTLEELK GI A +IGI+VD RR+N+ ES N+ RLK Y SKL+LF
Sbjct 68 KLRAGRGFTLEELKGAGIGANFAKTIGIAVDRRRKNKSLESRQRNIQRLKEYRSKLILFP 127
Query 136 RGSKAKKGLAGVPADTPKSQLQNVKQAK--IADSLPI--ERPRKKIRARVITEEERKFRA 191
KK AG +S L+ K A LPI E+P + R +T++E+KF+A
Sbjct 128 INE--KKIRAG------ESSLEECKLATQLKGPVLPIKNEQP-AVVEFREVTKDEKKFKA 178
Query 192 YATLRKQLRDAKNVGKHAKKLAEEAEAAN 220
+ATLRK DA+ VG AK+ E AE+ +
Sbjct 179 FATLRKARTDARLVGIRAKRAKEAAESED 207
> Hs15431295
Length=211
Score = 161 bits (407), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 103/208 (49%), Positives = 132/208 (63%), Gaps = 9/208 (4%)
Query 12 SRSAQLPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPL-GLLRPAVRCPT 70
SR+ + H K+WQR V TWF+QPARK RRR RQ KA ++ RP G +RP VRCPT
Sbjct 4 SRNGMVLKPHFHKDWQRRVATWFNQPARKIRRRKARQAKARRIAPRPASGPIRPIVRCPT 63
Query 71 QKYNIKLREGRGFTLEELKAVGISPRVALSIGISVDHRRRNRCQESLNTNVNRLKLYLSK 130
+Y+ K+R GRGF+LEEL+ GI +VA +IGISVD RRRN+ ESL NV RLK Y SK
Sbjct 64 VRYHTKVRAGRGFSLEELRVAGIHKKVARTIGISVDPRRRNKSTESLQANVQRLKEYRSK 123
Query 131 LVLFQRGSKA-KKGLAGVPADTPKSQLQNVKQAKIADSLPIERPRKKIRARVITEEERKF 189
L+LF R A KKG D+ +L+ Q +P+ KK +ARVITEEE+ F
Sbjct 124 LILFPRKPSAPKKG------DSSAEELKLATQL-TGPVMPVRNVYKKEKARVITEEEKNF 176
Query 190 RAYATLRKQLRDAKNVGKHAKKLAEEAE 217
+A+A+LR +A+ G AK+ E AE
Sbjct 177 KAFASLRMARANARLFGIRAKRAKEAAE 204
> CE08526
Length=207
Score = 158 bits (399), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 97/211 (45%), Positives = 125/211 (59%), Gaps = 10/211 (4%)
Query 10 MVSRSAQ-LPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPL-GLLRPAVR 67
M R Q L N H RK W + ++TWFDQPARK RRR RQ KA ++ RP+ GLLR VR
Sbjct 1 MAPRGNQMLGNAHFRKHWHKRIKTWFDQPARKLRRRQNRQAKAVEIAPRPVAGLLRSVVR 60
Query 68 CPTQKYNIKLREGRGFTLEELKAVGISPRVALSIGISVDHRRRNRCQESLNTNVNRLKLY 127
CP ++YN K R GRGF+L+ELKA GIS A +IGI+VD RR N+ E L N +RLK Y
Sbjct 61 CPQKRYNTKTRLGRGFSLQELKAAGISQAQARTIGIAVDVRRTNKTAEGLKANADRLKEY 120
Query 128 LSKLVLF-QRGSKAKKGLAGVPADTPKSQLQNVKQAKIADSLPIERPRKKIRARVITEEE 186
+KL+LF ++ S KKG D+ +L+ Q + D LP+ R +T+ E
Sbjct 121 KAKLILFPKKASAPKKG------DSSAEELKVAAQLR-GDVLPLSHTITFDEPRQVTDAE 173
Query 187 RKFRAYATLRKQLRDAKNVGKHAKKLAEEAE 217
RK + LRK+ D K GK K+ E AE
Sbjct 174 RKVEIFRLLRKERADKKYRGKREKRAREAAE 204
> At3g48960
Length=206
Score = 153 bits (386), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 82/191 (42%), Positives = 118/191 (61%), Gaps = 7/191 (3%)
Query 17 LPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPL-GLLRPAVRCPTQKYNI 75
+PN H +K+W+ +V+T F+QPA K RRR+ RQ KA K+ RP G +RP V T YN+
Sbjct 7 IPNGHFKKKWENYVKTSFNQPAMKTRRRIARQNKAVKIFPRPTAGPIRPVVHAQTLTYNM 66
Query 76 KLREGRGFTLEELKAVGISPRVALSIGISVDHRRRNRCQESLNTNVNRLKLYLSKLVLFQ 135
K+R G+GFTLEELKA GI ++A +IGISVDH R+NR E TNV RLK Y +KLV+F
Sbjct 67 KVRAGKGFTLEELKAAGIPKKLAPTIGISVDHHRKNRSLEGFQTNVQRLKTYKAKLVIFP 126
Query 136 RGSKAKKGLAGVPADTPKSQLQNVKQAKIADSLPIERPRKKIRARVITEEERKFRAYATL 195
R ++ K D+ + +L N Q ++ D +PI R + +T + + F AY +
Sbjct 127 RCARTVK-----VGDSAQQELANATQVQV-DHMPIVREMPTMELVKLTSDMKLFNAYDKI 180
Query 196 RKQLRDAKNVG 206
R + + ++ G
Sbjct 181 RLEGINKRHAG 191
> YDL082w
Length=199
Score = 129 bits (323), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 79/186 (42%), Positives = 106/186 (56%), Gaps = 17/186 (9%)
Query 11 VSRSAQLPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPLGLLRPAVRCPT 70
+S++ + H RK WQ V+ FDQ +K RR R +AAK+ RPL LLRP VR PT
Sbjct 3 ISKNLPILKNHFRKHWQERVKVHFDQAGKKVSRRNARATRAAKIAPRPLDLLRPVVRAPT 62
Query 71 QKYNIKLREGRGFTLEELKAVGISPRVALSIGISVDHRRRNRCQESLNTNVNRLKLYLSK 130
KYN K+R GRGFTL E+KA G++ A +IGI+VDHRR+NR QE + NV RLK Y SK
Sbjct 63 VKYNRKVRAGRGFTLAEVKAAGLTAAYARTIGIAVDHRRQNRNQEIFDANVQRLKEYQSK 122
Query 131 LVLFQRGSKAKKGLAGVPADTPKSQLQNVKQAKIADSLPIERPRKKIRARVITEEERKFR 190
+++F R KA + + A A + PI +P + AR + +
Sbjct 123 IIVFPRNGKAPEAEQVLSA---------------AATFPIAQPATDVEARAVQDNGES-- 165
Query 191 AYATLR 196
A+ TLR
Sbjct 166 AFRTLR 171
> YMR142c
Length=199
Score = 123 bits (309), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 79/186 (42%), Positives = 106/186 (56%), Gaps = 17/186 (9%)
Query 11 VSRSAQLPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPLGLLRPAVRCPT 70
+S++ + H RK WQ V+ FDQ +K RR R +AAK+ RPL LLRP VR PT
Sbjct 3 ISKNLPILKNHFRKHWQERVKVHFDQAGKKVSRRNARAARAAKIAPRPLDLLRPVVRAPT 62
Query 71 QKYNIKLREGRGFTLEELKAVGISPRVALSIGISVDHRRRNRCQESLNTNVNRLKLYLSK 130
KYN K+R GRGFTL E+KA G++ A +IGI+VDHRR+NR QE + NV RLK Y SK
Sbjct 63 VKYNRKVRAGRGFTLAEVKAAGLTAAYARTIGIAVDHRRQNRNQEIFDANVQRLKEYQSK 122
Query 131 LVLFQRGSKAKKGLAGVPADTPKSQLQNVKQAKIADSLPIERPRKKIRARVITEEERKFR 190
+++F R KA + + A A + PI +P + AR + +
Sbjct 123 IIVFPRDGKAPEAEQVLSA---------------AATFPIAQPATDVEARAVQDNGES-- 165
Query 191 AYATLR 196
A+ TLR
Sbjct 166 AFRTLR 171
> ECU03g0320
Length=163
Score = 89.0 bits (219), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 56/155 (36%), Positives = 85/155 (54%), Gaps = 7/155 (4%)
Query 17 LPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPLGLLRPAVRCPTQKYNIK 76
LPN H RK + +R D + + ++ +KA L PL LRP VRCPT KYN
Sbjct 7 LPNNHFRKTSLK-IRIHHDPETKARVMAEKKLRKAKALFPMPLKKLRPIVRCPTIKYNRN 65
Query 77 LREGRGFTLEELKAVGISPRVALSIGISVDHRRRNRCQESLNTNVNRLKLYLSKLVLFQR 136
R GRGFT E + G+ R A +GI+VD RRR+ QE+ + NV R+K YL K+ +++
Sbjct 66 ERLGRGFTAAECEKAGLDYRHARRLGIAVDLRRRDTNQEAFDKNVERIKTYLGKITIYES 125
Query 137 GSKAKKGLAG------VPADTPKSQLQNVKQAKIA 165
+A++ A +P PK + ++ + ++A
Sbjct 126 VKEARESGAKPYTKEIMPFVKPKPVVGSISREEVA 160
> Hs22045017
Length=180
Score = 84.7 bits (208), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 60/134 (44%), Positives = 74/134 (55%), Gaps = 8/134 (5%)
Query 85 LEELKAVGISPRVALSIGISVDHRRRNRCQESLNTNVNRLKLYLSKLVLFQRGSKA-KKG 143
LEELK GI VA + GIS+D RRRN+ ESL NV RLK Y SKL+LF R A KKG
Sbjct 43 LEELKVAGIHKNVAQTTGISIDWRRRNKSTESLQGNVQRLKEYNSKLILFPRKPLAPKKG 102
Query 144 LAGVPADTPKSQLQNVKQAKIADSLPIERPRKKIRARVITEEERKFRAYATLRKQLRDAK 203
+ V QL +PI++ KK +A VITEEE+ F+A+A L A+
Sbjct 103 DSSVEELILAIQL-------TGPVMPIQKVYKKEKALVITEEEKNFKAFAHLLMACAIAQ 155
Query 204 NVGKHAKKLAEEAE 217
G AK+ E E
Sbjct 156 LFGIKAKRAKEAVE 169
> Hs14784147
Length=144
Score = 73.2 bits (178), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 49/121 (40%), Positives = 68/121 (56%), Gaps = 7/121 (5%)
Query 64 PAVRCPTQKYNIKLREGRGFTLEELKAVGISPRVALSIGISVDHRRRNRCQESLNTNVNR 123
P VRCP +++ K + GRG +LEEL+ GI +VA +IGIS D RRRN+ ++L V R
Sbjct 31 PIVRCPMLRHHYKAQAGRGLSLEELRVAGIYKKVAQTIGISEDARRRNQSTQALQAKVQR 90
Query 124 LKLYLSKLVLFQRGSKAKKGLAGVPADTPKSQLQNVKQAKIADSLPIERPRKKIRARVIT 183
LK S L+LF R K LA D+ +L+ Q +PI K+ +ARVI
Sbjct 91 LKEDRSSLILFPR-----KPLAPKKGDSSAEELELDTQL-TGPEMPIGNVYKE-KARVIA 143
Query 184 E 184
+
Sbjct 144 D 144
> Hs17466264
Length=161
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 35/83 (42%), Gaps = 24/83 (28%)
Query 12 SRSAQLPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPLGLLRPA-VRCPT 70
SR + H K WQ V TWF+ PA +RC
Sbjct 4 SRKGMIFKSHFNKHWQWCVATWFNHV-----------------------WTHPAHIRCCM 40
Query 71 QKYNIKLREGRGFTLEELKAVGI 93
+Y+ K+ G+GF++EEL+ VGI
Sbjct 41 VRYHTKVHAGQGFSMEELRVVGI 63
> Hs22070432
Length=119
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 19/33 (57%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 12 SRSAQLPNVHLRKEWQRFVRTWFDQPARKQRRR 44
SR+ + H K+WQR V TWF+QPARK RRR
Sbjct 4 SRNGMVLKPHFHKDWQRRVATWFNQPARKIRRR 36
> Hs17440716
Length=245
Score = 30.8 bits (68), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 20/75 (26%), Positives = 35/75 (46%), Gaps = 8/75 (10%)
Query 2 CCCFPQAKMVSRSAQLPNVHLRKEWQRFVRTWFDQPARKQRRRLQRQKKAAKLGVRPLG- 60
C P+++ +S++A LPN+ WQ + D AR + ++ ++ +R G
Sbjct 40 CEVKPESQHISKTAHLPNLSNEIRWQDSMEVGSDSEARSEMNIIKPCSMTQRVEIRLCGH 99
Query 61 -----LLRP--AVRC 68
LL+P AV C
Sbjct 100 ALLSKLLQPPEAVNC 114
> Hs21450713
Length=787
Score = 30.4 bits (67), Expect = 2.6, Method: Composition-based stats.
Identities = 21/60 (35%), Positives = 31/60 (51%), Gaps = 1/60 (1%)
Query 57 RPLGLLRPAVRCPTQKYNIKLREGRGFTLEELKAVGISPRVALSIGISVDHRRRNRCQES 116
R LG + P C T+ + TLE AV ++ +ALS+GIS D ++ +C ES
Sbjct 284 RYLGAVFPGTMCITRYSAGVALYPKEITLEAF-AVIVTQMLALSLGISYDDPKKCQCSES 342
> ECU10g0800
Length=210
Score = 29.6 bits (65), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 35/71 (49%), Gaps = 4/71 (5%)
Query 41 QRRRLQRQKKAAKLGVRPLGLLRPAVRCPTQKYNIKLREGRGFTLEELK----AVGISPR 96
RRR+ + + V L ++ + PT++ N LR G + + E++ +VG+ R
Sbjct 103 MRRRIPLDDGSVDVAVCCLSMMVEDIAVPTKEINRILRNGGHWYIAEVRSRIPSVGLLER 162
Query 97 VALSIGISVDH 107
S+G V+H
Sbjct 163 KFESLGFDVEH 173
> YER171w
Length=778
Score = 29.3 bits (64), Expect = 6.3, Method: Composition-based stats.
Identities = 22/76 (28%), Positives = 38/76 (50%), Gaps = 4/76 (5%)
Query 115 ESLNTNVNRLKLYLSKLVLFQRGSKAKKGLAGVPADTPKSQLQNVKQAKIADSLPI---- 170
E++ N+ R + ++S L K + + V ++TPKS LQ++KQ + P+
Sbjct 319 EAIPGNIRRAEHFVSFLKRLIEYLKTRMKVLHVISETPKSFLQHLKQLTFIERKPLRFCS 378
Query 171 ERPRKKIRARVITEEE 186
ER +R +TE E
Sbjct 379 ERLSLLVRTLEVTEVE 394
> 7303535
Length=1137
Score = 28.9 bits (63), Expect = 9.1, Method: Composition-based stats.
Identities = 29/118 (24%), Positives = 52/118 (44%), Gaps = 3/118 (2%)
Query 88 LKAVGISPRVALSIGISVDHRRRNRCQESLNTNVNRLKLYLSKLVLFQRGSK-AKKGLAG 146
KA+G+ + I +D + +NR SL N LK++ + L GSK K
Sbjct 247 FKALGVVSDQEIQSLIGIDSKSQNRFGASLIDAYN-LKVFTQQRALEYMGSKLVVKRFQS 305
Query 147 VPADTPKSQLQNVKQAKIADSLPIERPRKKIRARVITEEERKFRAYATLRKQLRDAKN 204
TP + + + I +P++ +++A ++ R+ A A L K L D ++
Sbjct 306 ATTKTPSEEARELLLTTILAHVPVDNFNLQMKAIYVSMMVRRVMA-AELDKTLFDDRD 362
Lambda K H
0.320 0.133 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4183546302
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40