bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3108_orf1
Length=437
Score E
Sequences producing significant alignments: (Bits) Value
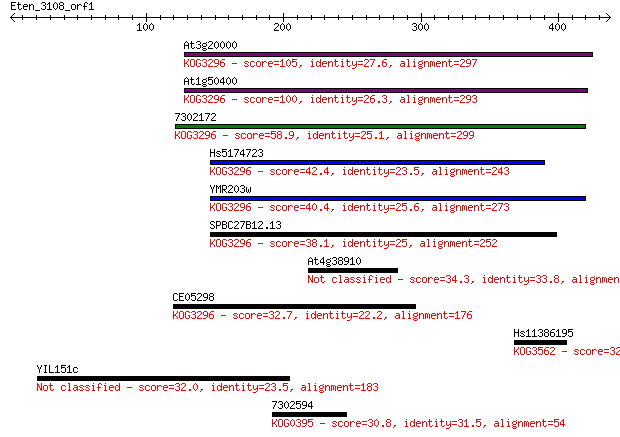
At3g20000 105 2e-22
At1g50400 100 9e-21
7302172 58.9 2e-08
Hs5174723 42.4 0.002
YMR203w 40.4 0.009
SPBC27B12.13 38.1 0.035
At4g38910 34.3 0.52
CE05298 32.7 1.5
Hs11386195 32.7 1.8
YIL151c 32.0 2.9
7302594 30.8 5.5
> At3g20000
Length=309
Score = 105 bits (262), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 82/311 (26%), Positives = 136/311 (43%), Gaps = 47/311 (15%)
Query 128 PIPFEHFSREWMNISGQDTFDGFRLEGAKQVNKYLQTCHSFCLG----------TQLRES 177
P+P+E RE + D F+G R + + +N+ HS +G T ++
Sbjct 31 PVPYEELHREALMSLKSDNFEGLRFDFTRALNQKFSLSHSVMMGPTEVPAQSPETTIKIP 90
Query 178 GYSYQFGPTVAISPPPDPSQGENAQPPPPHFFAMAKLGTDGSLQGRLIKTLCQSADLKLN 237
Y+FG A P + ++ TDG L RL L +K N
Sbjct 91 TAHYEFG----------------ANYYDPKLLLIGRVMTDGRLNARLKADLTDKLVVKAN 134
Query 238 FNSSLKDEQRNFYEISLDNTGRDWASSLKVAWQSCWILDGMFSQVLTPRLQAGCEITYVG 297
+ +E + + D G D+ + L++ QS I + Q +T L G EI + G
Sbjct 135 ALIT-NEEHMSQAMFNFDYMGSDYRAQLQLG-QSALI-GATYIQSVTNHLSLGGEIFWAG 191
Query 298 ANGASMLAVGGRYNLDKQSVVSCQLSQQPDFKSPAGLTKLTHTCKVQYARKVTDRLSMAS 357
S + RY DK V S Q++ T + Y +K++D++S+A+
Sbjct 192 VPRKSGIGYAARYETDKM-VASGQVAS-------------TGAVVMNYVQKISDKVSLAT 237
Query 358 EFEYSHPETESALRVGWEYLFRQARVQGLIDSCGRISVFAQDYNGFGVSGL----IDYWR 413
+F Y++ + VG++Y+ RQARV+G IDS G S ++ G++ L +D+ +
Sbjct 238 DFMYNYFSRDVTASVGYDYMLRQARVRGKIDSNGVASALLEERLSMGLNFLLSAELDHKK 297
Query 414 GTYKFGFLMHV 424
YKFGF + V
Sbjct 298 KDYKFGFGLTV 308
> At1g50400
Length=310
Score = 100 bits (248), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 77/299 (25%), Positives = 141/299 (47%), Gaps = 30/299 (10%)
Query 128 PIPFEHFSREWMNISGQDTFDGFRLEGAKQVNKYLQTCHSFCLG-TQLRESGYSYQFGPT 186
P+ +E +RE + F+GFRL+ K +N+ HS +G T++ P+
Sbjct 31 PVLYEELNREATMALKPELFEGFRLDYNKSLNQKFFLSHSILMGPTEVPNPT------PS 84
Query 187 VAISPPPDPSQGENAQPPPPHFFAMAKLGTDGSLQGRLIKTLCQSADLKLNFNSSLKDEQ 246
I P + A P + + ++ TDG L R L + +K N + L DE+
Sbjct 85 SEIIKIPTANYDFGAGFIDPKLYLIGRITTDGRLNARAKFDLTDNFSVKAN--ALLTDEE 142
Query 247 -RNFYEISLDNTGRDWASSLKVAWQSCWILDGMFSQVLTPRLQAGCEITYVGANGASMLA 305
++ + +D G D+ + L++ S + + + Q +TP L G E ++G S +
Sbjct 143 DKSQGHLVIDYKGSDYRTQLQLGNNSVYAAN--YIQHVTPHLSLGGEAFWLGQQLMSGVG 200
Query 306 VGGRYNLDKQSVVSCQLSQQPDFKSPAGLTKLTHTCKVQYARKVTDRLSMASEFEYSHPE 365
RY DK +V S Q++ T + Y KV+++LS A++F Y++
Sbjct 201 YAARYETDK-TVASGQIAS-------------TGVAVMNYVHKVSEKLSFATDFIYNYLS 246
Query 366 TESALRVGWEYLFRQARVQGLIDSCGRISVFAQDYNGFG----VSGLIDYWRGTYKFGF 420
+ VG++ + RQ+R++G +DS G ++ + ++ G +S +D+ + YKFGF
Sbjct 247 RDVTASVGYDLITRQSRLRGKVDSNGVVAAYLEEQLPIGLRFLLSAEVDHVKKDYKFGF 305
> 7302172
Length=340
Score = 58.9 bits (141), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 75/316 (23%), Positives = 136/316 (43%), Gaps = 47/316 (14%)
Query 121 KKPQPTQPIPFEHFSREWMNISGQDTFDGFRLEGAKQVNKYLQTCHSFCLGTQLRESGYS 180
KKP+ T P E +I +TF+G ++ K ++ + Q H+ + +G S
Sbjct 49 KKPELTNPGTLEELHSRCRDIQA-NTFEGAKIMVNKGLSNHFQVTHTI----NMNSAGPS 103
Query 181 -YQFGPTVAISPPPDPSQGENAQPPPPHFFAMAKLGTDGSLQGRLIKTLCQSADLKLNFN 239
Y+FG T + P++ A P + ++ G+L +I L ++ L+ F
Sbjct 104 GYRFGATYVGTKQYGPTE---AFP-----VLLGEIDPMGNLNANVIHQL--TSRLRCKFA 153
Query 240 SSLKDEQRNFYEISLDNTGRDWASSLKVAWQSCWILDGMFS----QVLTPRLQAGCEITY 295
S +D + +++ D GRD+ +L + + G+F Q +T RL G E Y
Sbjct 154 SQFQDSKLVGTQLTGDYRGRDYTLTLTMGNPGFFTSSGVFVCQYLQSVTKRLALGSEFAY 213
Query 296 -----VGANGASMLAVGGRYNLDKQSVVSCQLSQQPDFKSPAGLTKLTHTCKVQYARKVT 350
V ++L+ GRY +V SC L PAG + Y +K +
Sbjct 214 HYGPNVPGRQVAVLSAVGRYAFG-DTVWSCTL-------GPAGF-------HLSYYQKAS 258
Query 351 DRLSMASEFEYSHPETESALRVGWEYLFRQARV--QGLIDSCGRIS-VFAQDYN----GF 403
D+L + E E + + ES V ++ +A + +G +DS IS V +
Sbjct 259 DQLQIGVEVETNIRQQESTATVAYQIDLPKADLVFRGSLDSNWLISGVLEKRLQPLPFSL 318
Query 404 GVSGLIDYWRGTYKFG 419
+SG +++ + +++ G
Sbjct 319 AISGRMNHQKNSFRLG 334
> Hs5174723
Length=361
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 57/253 (22%), Positives = 109/253 (43%), Gaps = 40/253 (15%)
Query 147 FDGFRLEGAKQVNKYLQTCHSFCLGTQLRESGYSYQFGPTVAISPPPDPSQGENAQPPPP 206
+G +L K ++ + Q H+ L T + ES +Y FG T + P++ A P
Sbjct 98 MEGVKLTVNKGLSNHFQVNHTVALST-IGES--NYHFGVTYVGTKQLSPTE---AFP--- 148
Query 207 HFFAMAKLGTDGSLQGRLIKTLCQSADLKLNFNSSLKDEQRNFYEISLDNT--GRDWASS 264
+ + GSL ++I L K+ +++ +Q F +D G D+ ++
Sbjct 149 --VLVGDMDNSGSLNAQVIHQLGPGLRSKM----AIQTQQSKFVNWQVDGEYRGSDFTAA 202
Query 265 LKV----AWQSCWILDGMFSQVLTPRLQAGCEITYVGANG--ASMLAVGGRYNLDKQSVV 318
+ + IL + Q +TP L G E+ Y G +++++ G+Y L+ +
Sbjct 203 VTLGNPDVLVGSGILVAHYLQSITPCLALGGELVYHRRPGEEGTVMSLAGKYTLNNW-LA 261
Query 319 SCQLSQQPDFKSPAGLTKLTHTCKVQYARKVTDRLSMASEFEYSHPETESALRVGWEYLF 378
+ L Q AG+ Y K +D+L + EFE S ++++ G++
Sbjct 262 TVTLGQ-------AGM-------HATYYHKASDQLQVGVEFEASTRMQDTSVSFGYQLDL 307
Query 379 RQARV--QGLIDS 389
+A + +G +DS
Sbjct 308 PKANLLFKGSVDS 320
> YMR203w
Length=387
Score = 40.4 bits (93), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 70/298 (23%), Positives = 113/298 (37%), Gaps = 49/298 (16%)
Query 147 FDGFR--LEGAKQVNKYLQTCHSFCLGTQ-LRESGYSYQFGPTVAISPPPDPSQGENAQP 203
F G R L A +N QT H+F +G+Q L + +S F
Sbjct 81 FTGLRADLNKAFSMNPAFQTSHTFSIGSQALPKYAFSALFAND----------------- 123
Query 204 PPPHFFAMAKLGTDGSLQGRLIKTLCQSADLKLNFNSSLKDEQRNFYEISLDNTGRDWAS 263
+ FA + D S+ GRL + K+N + D Q ++ D D++
Sbjct 124 ---NLFAQGNIDNDLSVSGRLNYGWDKKNISKVNLQ--ISDGQPTMCQLEQDYQASDFSV 178
Query 264 SLKVAWQS-------CWILDGMFSQVLTPRLQAGCEITYVGANGASMLAVG----GRYNL 312
++K S + F Q +TP+L G E Y +G++ G RY
Sbjct 179 NVKTLNPSFSEKGEFTGVAVASFLQSVTPQLALGLETLYSRTDGSAPGDAGVSYLTRYVS 238
Query 313 DKQS-VVSCQLSQQPDF------KSPAGLTKLTHTCKVQYARKVTDRLSMASEFEYSHPE 365
KQ + S QL K + T +TD L M + P
Sbjct 239 KKQDWIFSGQLQANGALIASLWRKVAQNVEAGIETTLQAGMVPITDPL-MGTPIGI-QPT 296
Query 366 TESALRVGWEYLFRQARVQGLIDSCGRISVFAQDYNGFGVS----GLIDYWRGTYKFG 419
E + +G +Y +RQ+ +G +DS G+++ F + +S G ID+++ K G
Sbjct 297 VEGSTTIGAKYEYRQSVYRGTLDSNGKVACFLERKVLPTLSVLFCGEIDHFKNDTKIG 354
> SPBC27B12.13
Length=309
Score = 38.1 bits (87), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 63/279 (22%), Positives = 105/279 (37%), Gaps = 63/279 (22%)
Query 147 FDGFRLEGAKQ--VNKYLQTCHSFCLGTQLRESGYSYQFGPTVAISPPPDPSQGENAQPP 204
F G R + K + + H+F LG+Q + PP S +P
Sbjct 51 FTGVRADVTKGFCTSPWFTVSHAFALGSQ---------------VLPPYSFSTMFGGEP- 94
Query 205 PPHFFAMAKLGTDGSLQGRLIKTLCQSADLKLNFNSSLKDEQRNFYEISLDNTGRDWASS 264
F + DG++Q L T + K+ S N +I D+ G+D++ S
Sbjct 95 ---LFLRGSVDNDGAVQAMLNCTWNSNVLSKVQMQLS-NGAVPNMCQIEHDHKGKDFSFS 150
Query 265 LKVAWQSCWI---LDGMFS----QVLTPRLQAGCEITY------VGANGASMLAVGGRYN 311
K + W L G++ Q +TP+L G E + +G A+ L+ RYN
Sbjct 151 FKA--MNPWYEEKLTGIYIISLLQSVTPKLSLGVEALWQKPSSSIGPEEAT-LSYMTRYN 207
Query 312 LDKQSVVSCQLSQQPDFKSPAGLTKLTHTCKVQYARKVTDRLSMASEFEYS--------- 362
D+ + A L + RK++ ++ E + S
Sbjct 208 -------------AADWIATAHLNGSQGDVTATFWRKLSPKVEAGVECQLSPVGLNHSAA 254
Query 363 ---HPETESALRVGWEYLFRQARVQGLIDSCGRISVFAQ 398
P+ E VG +Y F Q+ +G +DS GR+ V+ +
Sbjct 255 LMTGPKPEGLTSVGVKYEFAQSIYRGQVDSKGRVGVYLE 293
> At4g38910
Length=445
Score = 34.3 bits (77), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 34/66 (51%), Gaps = 4/66 (6%)
Query 218 GSLQGRLIKTLCQSADLKLNFNSSLKDEQRNFYEISLDNT-GRDWASSLKVAWQSCWILD 276
GS+ RL+ L ADL + S +KDE R ++D++ R +SS + W C+I
Sbjct 258 GSVFSRLLSRL---ADLTIRIKSRMKDEVRLVIVFTVDSSVKRQISSSTVLTWFCCFIFA 314
Query 277 GMFSQV 282
FS +
Sbjct 315 TFFSGI 320
> CE05298
Length=301
Score = 32.7 bits (73), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 39/187 (20%), Positives = 69/187 (36%), Gaps = 28/187 (14%)
Query 120 PKKPQPTQPIP------FEHFSREWMNISGQDTFDGFRLEGAKQVNKYLQTCHSFCLGTQ 173
P + + PIP +E R+ ++ F+G +L K ++ + Q H+ L
Sbjct 4 PTESELASPIPQTNPGSYEELHRKARDVF-PTCFEGAKLMVNKGLSSHFQVSHTLSLSAM 62
Query 174 LRESGYSYQFGPTVAISPPPDPSQGENAQPPPPHFFAMAKLGTDGSLQGRLIKTLCQSAD 233
Y+FG T + P++ A L D + G T+
Sbjct 63 ----NTGYRFGATYVGTNQVGPAE------------AYPILLGDTDVNGNTTATILHQLG 106
Query 234 L-KLNFNSSLKDEQRNFYEISLDNTGRDWASSLKVA----WQSCWILDGMFSQVLTPRLQ 288
+ + ++ + + +++ GR L +A IL G F + LTPRL
Sbjct 107 IYRTKLQGQIQQGKLAGAQATIERKGRLSTLGLTLANIDLVNEAGILVGQFLRRLTPRLD 166
Query 289 AGCEITY 295
G E+ Y
Sbjct 167 VGTEMVY 173
> Hs11386195
Length=324
Score = 32.7 bits (73), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 24/38 (63%), Gaps = 3/38 (7%)
Query 368 SALRVGWEYLFRQARVQGLIDSCGRISVFAQDYNGFGV 405
S+L GW +++ A G+IDS IS+ + DY+ +GV
Sbjct 208 SSLGTGWPHVYGSA---GMIDSATPISMASGDYSAYGV 242
> YIL151c
Length=1118
Score = 32.0 bits (71), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 43/192 (22%), Positives = 78/192 (40%), Gaps = 14/192 (7%)
Query 21 DFLRRGDQRADGCRQRRGESVRGGRSRCASSVSVGKDDSPAVARCSEKSSGFLGVPGLGF 80
D+ R + R S + SS S +D+P + + G + G GF
Sbjct 17 DYSRSNNNHTHIVSDMRPTSAAFLHQKRHSSSS--HNDTPESSFAKRRVPGIVDPVGKGF 74
Query 81 FGGSKESQSASSDAPAAVDKREKARLDTLARE-NEQTPEPPKKPQPTQPIPFE--HFSRE 137
G SQ ++ + P+ D + +R +++R+ E TP+ PT +P + +R
Sbjct 75 IDGITNSQISAQNTPSKTD--DASRRPSISRKVMESTPQVKTSSIPTMDVPKSPYYVNRT 132
Query 138 W----MNISGQDTFDGFRLEGAKQVNKYLQTCHSFCLGTQLRE--SGYSYQFGPTVAISP 191
M + +DT++ + ++ L + +Q + + + P VA SP
Sbjct 133 MLARNMKVVSRDTYED-NANPQMRADEPLVASNGIYSNSQPQSQVTLSDIRRAPVVAASP 191
Query 192 PPDPSQGENAQP 203
PP Q +AQP
Sbjct 192 PPMIRQLPSAQP 203
> 7302594
Length=740
Score = 30.8 bits (68), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 25/54 (46%), Gaps = 1/54 (1%)
Query 192 PPDPSQGENAQPPPPHFFAMAKLGTDGSLQGRLIKTLCQSADLKLNFNSSLKDE 245
P P EN PPPP + + LG G + L+ S + +++SL DE
Sbjct 510 PATPDDSENLDPPPPARYRVVMLGDAGVGKTALVNQFMTSEYMH-TYDASLDDE 562
Lambda K H
0.317 0.134 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 11327369950
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40