bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3091_orf1
Length=137
Score E
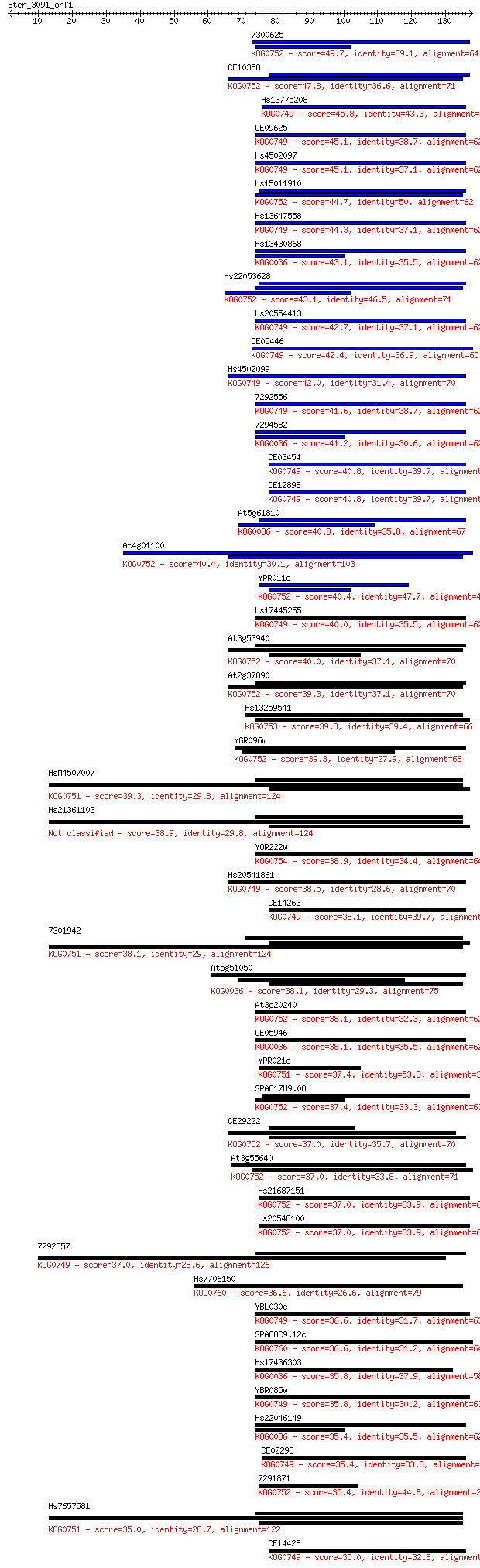
Sequences producing significant alignments: (Bits) Value
7300625 49.7 2e-06
CE10358 47.8 7e-06
Hs13775208 45.8 2e-05
CE09625 45.1 4e-05
Hs4502097 45.1 4e-05
Hs15011910 44.7 5e-05
Hs13647558 44.3 6e-05
Hs13430868 43.1 2e-04
Hs22053628 43.1 2e-04
Hs20554413 42.7 2e-04
CE05446 42.4 2e-04
Hs4502099 42.0 4e-04
7292556 41.6 5e-04
7294582 41.2 6e-04
CE03454 40.8 7e-04
CE12898 40.8 7e-04
At5g61810 40.8 8e-04
At4g01100 40.4 0.001
YPR011c 40.4 0.001
Hs17445255 40.0 0.001
At3g53940 40.0 0.001
At2g37890 39.3 0.002
Hs13259541 39.3 0.002
YGR096w 39.3 0.002
HsM4507007 39.3 0.002
Hs21361103 38.9 0.003
YOR222w 38.9 0.003
Hs20541861 38.5 0.004
CE14263 38.1 0.005
7301942 38.1 0.005
At5g51050 38.1 0.005
At3g20240 38.1 0.005
CE05946 38.1 0.006
YPR021c 37.4 0.009
SPAC17H9.08 37.4 0.009
CE29222 37.0 0.010
At3g55640 37.0 0.011
Hs21687151 37.0 0.011
Hs20548100 37.0 0.011
7292557 37.0 0.011
Hs7706150 36.6 0.014
YBL030c 36.6 0.015
SPAC8C9.12c 36.6 0.016
Hs17436303 35.8 0.024
YBR085w 35.8 0.026
Hs22046149 35.4 0.035
CE02298 35.4 0.036
7291871 35.4 0.036
Hs7657581 35.0 0.041
CE14428 35.0 0.041
> 7300625
Length=377
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/64 (39%), Positives = 35/64 (54%), Gaps = 3/64 (4%)
Query 73 CQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSLW 132
+ G+ AG +AKTV+ PLDR K+ Q+++ FS R R LQ G +LW
Sbjct 86 ISLISGAAAGALAKTVIAPLDRTKINFQIRN---DVPFSFRASLRYLQNTYANEGVLALW 142
Query 133 KGNS 136
+GNS
Sbjct 143 RGNS 146
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 11/28 (39%), Positives = 20/28 (71%), Gaps = 0/28 (0%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQV 101
+FL GS+AG ++++ YPLD + ++ V
Sbjct 183 RFLAGSLAGITSQSLTYPLDLARARMAV 210
> CE10358
Length=294
Score = 47.8 bits (112), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 24/59 (40%), Positives = 37/59 (62%), Gaps = 3/59 (5%)
Query 78 GSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSLWKGNS 136
G+IAG +AKT + PLDR K+ QV S + +S R + ++ + GFF+L++GNS
Sbjct 21 GAIAGALAKTTIAPLDRTKIYFQVSS---TRGYSFRSAIKFIKLTYRENGFFALYRGNS 76
Score = 36.6 bits (83), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 27/72 (37%), Positives = 37/72 (51%), Gaps = 11/72 (15%)
Query 66 NTSICATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQT 125
N S +++ GS+A T A + YPLD K +L V S S SL+HV FV+T
Sbjct 105 NGSRTPVKRYITGSLAATTATMITYPLDTAKARLSVSSKLQYS--SLKHV------FVKT 156
Query 126 ---GGFFSLWKG 134
GG L++G
Sbjct 157 YKEGGIQLLYRG 168
> Hs13775208
Length=315
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 38/69 (55%), Gaps = 16/69 (23%)
Query 76 LRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRH---------VPRELQTFVQTG 126
L G +A ++KT V P++R+K+ LQVQ++S S R+ +PRE
Sbjct 25 LAGGVAAAVSKTAVAPIERVKLLLQVQASSKQISPEARYKGMVDCLVRIPRE-------Q 77
Query 127 GFFSLWKGN 135
GFFS W+GN
Sbjct 78 GFFSFWRGN 86
> CE09625
Length=326
Score = 45.1 bits (105), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 39/71 (54%), Gaps = 16/71 (22%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRH---------VPRELQTFVQ 124
F+ G+ A I+KTV+ P++R+K+ LQ+Q++ + + R+ VPRE
Sbjct 41 DFMAGATAAAISKTVIAPVERVKLILQLQNSQTTLALENRYKGIVDCFIRVPRE------ 94
Query 125 TGGFFSLWKGN 135
GF S W+GN
Sbjct 95 -QGFLSFWRGN 104
> Hs4502097
Length=297
Score = 45.1 bits (105), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 38/64 (59%), Gaps = 2/64 (3%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSAS--VSSSFSLRHVPRELQTFVQTGGFFSL 131
FL G++A ++KT V P++R+K+ LQVQ AS +S+ + + + + GF S
Sbjct 11 DFLAGAVAAAVSKTAVAPIERVKLLLQVQHASKQISAEKQYKGIIDCVVRIPKEQGFLSF 70
Query 132 WKGN 135
W+GN
Sbjct 71 WRGN 74
> Hs15011910
Length=332
Score = 44.7 bits (104), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 31/64 (48%), Positives = 35/64 (54%), Gaps = 10/64 (15%)
Query 75 FLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSS--SFS-LRHVPRELQTFVQTGGFFSL 131
FL GSIAG AKT V PLDR+K+ LQ + FS LR VP Q GF L
Sbjct 40 FLAGSIAGCCAKTTVAPLDRVKVLLQAHNHHYKHLGVFSALRAVP-------QKEGFLGL 92
Query 132 WKGN 135
+KGN
Sbjct 93 YKGN 96
Score = 28.5 bits (62), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 15/61 (24%), Positives = 30/61 (49%), Gaps = 2/61 (3%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSLWK 133
+ + GS+AG A P+D ++++L Q + H + + + + GGFF ++
Sbjct 133 RLMAGSMAGMTAVICTDPVDMVRVRLAFQVKGEHRYTGIIHAFKTI--YAKEGGFFGFYR 190
Query 134 G 134
G
Sbjct 191 G 191
> Hs13647558
Length=298
Score = 44.3 bits (103), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 37/64 (57%), Gaps = 2/64 (3%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSAS--VSSSFSLRHVPRELQTFVQTGGFFSL 131
FL G +A ++KT V P++R+K+ LQVQ AS +S+ + + + + GF S
Sbjct 11 DFLAGGVAAAVSKTAVAPIERVKLLLQVQHASKQISAEKQYKGIIDCVVRIPKEQGFLSF 70
Query 132 WKGN 135
W+GN
Sbjct 71 WRGN 74
> Hs13430868
Length=482
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 38/62 (61%), Gaps = 4/62 (6%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSLWK 133
Q + G++AG +++T PLDRLK+ +QV ++ + ++ L++ V GG SLW+
Sbjct 235 QLVAGAVAGAVSRTGTAPLDRLKVFMQVHASKTNR----LNILGGLRSMVLEGGIRSLWR 290
Query 134 GN 135
GN
Sbjct 291 GN 292
Score = 33.1 bits (74), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 21/26 (80%), Gaps = 0/26 (0%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKL 99
+F+ GS+AG A+T++YP++ LK +L
Sbjct 329 RFVAGSLAGATAQTIIYPMEVLKTRL 354
> Hs22053628
Length=332
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/64 (46%), Positives = 34/64 (53%), Gaps = 10/64 (15%)
Query 75 FLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSS--SFS-LRHVPRELQTFVQTGGFFSL 131
FL G IAG AKT V PLDR+K+ LQ + FS LR VP Q GF L
Sbjct 40 FLAGGIAGCCAKTTVAPLDRVKVLLQAHNHHYKHLGVFSALRAVP-------QKEGFLGL 92
Query 132 WKGN 135
+KGN
Sbjct 93 YKGN 96
Score = 36.2 bits (82), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 32/61 (52%), Gaps = 2/61 (3%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSLWK 133
+ + GS+AG A YPLD ++++L Q S + H + + + + GGFF ++
Sbjct 133 RLMAGSMAGMTAVICTYPLDMVRVRLAFQVKGEHSYTGIIHAFKTI--YAKEGGFFGFYR 190
Query 134 G 134
G
Sbjct 191 G 191
Score = 28.5 bits (62), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 13/37 (35%), Positives = 21/37 (56%), Gaps = 4/37 (10%)
Query 65 LNTSICATCQFLRGSIAGTIAKTVVYPLDRLKMKLQV 101
L T + C G +AG IA+T+ YP D + ++Q+
Sbjct 238 LKTHVNLLC----GGVAGAIAQTISYPFDVTRRRMQL 270
> Hs20554413
Length=298
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 36/64 (56%), Gaps = 2/64 (3%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSAS--VSSSFSLRHVPRELQTFVQTGGFFSL 131
FL G IA I+KT V P++R+K+ LQVQ AS +++ + + + + G S
Sbjct 11 DFLAGGIAAAISKTAVAPIERVKLLLQVQHASKQIAADKQYKGIVDCIVRIPKEQGVLSF 70
Query 132 WKGN 135
W+GN
Sbjct 71 WRGN 74
> CE05446
Length=306
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 40/74 (54%), Gaps = 16/74 (21%)
Query 73 CQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRE------LQTFVQT- 125
L G ++ +++KTVV P++R+K+ LQVQ +S + +P + + FV+
Sbjct 23 IDLLIGGVSASVSKTVVAPIERVKILLQVQ-------YSHKDIPADKRYNGIIDAFVRVP 75
Query 126 --GGFFSLWKGNST 137
GF S W+GN T
Sbjct 76 KEQGFVSFWRGNMT 89
> Hs4502099
Length=298
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 39/72 (54%), Gaps = 2/72 (2%)
Query 66 NTSICATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSAS--VSSSFSLRHVPRELQTFV 123
+ ++ FL G +A I+KT V P++R+K+ LQVQ AS +++ + + +
Sbjct 3 DAAVSFAKDFLAGGVAAAISKTAVAPIERVKLLLQVQHASKQITADKQYKGIIDCVVRIP 62
Query 124 QTGGFFSLWKGN 135
+ G S W+GN
Sbjct 63 KEQGVLSFWRGN 74
> 7292556
Length=307
Score = 41.6 bits (96), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 36/71 (50%), Gaps = 16/71 (22%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRH---------VPRELQTFVQ 124
F+ G ++ IAKT V P++R+K+ LQVQ S + R+ +P+E
Sbjct 21 DFMMGGVSAAIAKTAVAPIERVKLILQVQEVSKQIAADQRYKGIVDCFIRIPKE------ 74
Query 125 TGGFFSLWKGN 135
GF S W+GN
Sbjct 75 -QGFSSFWRGN 84
> 7294582
Length=370
Score = 41.2 bits (95), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 31/62 (50%), Gaps = 0/62 (0%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSLWK 133
+ G IAG +++T PLDR+K+ LQV + + + + GG S+W+
Sbjct 68 HLVAGGIAGAVSRTCTAPLDRIKVYLQVNQPRYTVQTQRMGISECMHIMLNEGGSRSMWR 127
Query 134 GN 135
GN
Sbjct 128 GN 129
Score = 30.0 bits (66), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 11/26 (42%), Positives = 20/26 (76%), Gaps = 0/26 (0%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKL 99
+F G+ AG I++T++YP++ LK +L
Sbjct 168 RFYAGAAAGGISQTIIYPMEVLKTRL 193
> CE03454
Length=313
Score = 40.8 bits (94), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 37/67 (55%), Gaps = 16/67 (23%)
Query 78 GSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRH---------VPRELQTFVQTGGF 128
G A ++KT V P++R+K+ LQVQ AS++ + R+ VP+E G+
Sbjct 32 GGTAAAVSKTAVAPIERVKLLLQVQDASLTIAADKRYKGIVDVLVRVPKE-------QGY 84
Query 129 FSLWKGN 135
+LW+GN
Sbjct 85 AALWRGN 91
> CE12898
Length=313
Score = 40.8 bits (94), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 37/67 (55%), Gaps = 16/67 (23%)
Query 78 GSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRH---------VPRELQTFVQTGGF 128
G A ++KT V P++R+K+ LQVQ AS++ + R+ VP+E G+
Sbjct 32 GGTAAAVSKTAVAPIERVKLLLQVQDASLTIAADKRYKGIVDVLVRVPKE-------QGY 84
Query 129 FSLWKGN 135
+LW+GN
Sbjct 85 AALWRGN 91
> At5g61810
Length=476
Score = 40.8 bits (94), Expect = 8e-04, Method: Composition-based stats.
Identities = 23/61 (37%), Positives = 35/61 (57%), Gaps = 7/61 (11%)
Query 75 FLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSLWKG 134
L G IAG +++T PLDRLK+ LQVQ ++ +++ + RE + GFF +G
Sbjct 208 LLAGGIAGAVSRTATAPLDRLKVALQVQRTNLGVVPTIKKIWREDKLL----GFF---RG 260
Query 135 N 135
N
Sbjct 261 N 261
Score = 34.3 bits (77), Expect = 0.067, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 69 ICATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSS 108
I + + L G +AG +A+T +YP+D +K +LQ + V +
Sbjct 293 IGTSGRLLAGGLAGAVAQTAIYPMDLVKTRLQTFVSEVGT 332
> At4g01100
Length=352
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 31/103 (30%), Positives = 51/103 (49%), Gaps = 5/103 (4%)
Query 35 KNRKRIKMTADEFRVSPQAAAAGGVEVSRHLNTSICATCQFLRGSIAGTIAKTVVYPLDR 94
++ KR + A V+ A GV+ + SIC + G +AG +++T V PL+R
Sbjct 4 EDVKRTESAAVSTIVNLAEEAREGVKAPSYAFKSICKS--LFAGGVAGGVSRTAVAPLER 61
Query 95 LKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSLWKGNST 137
+K+ LQVQ+ + + L+ +T G L+KGN T
Sbjct 62 MKILLQVQN---PHNIKYSGTVQGLKHIWRTEGLRGLFKGNGT 101
Score = 34.7 bits (78), Expect = 0.061, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 43/80 (53%), Gaps = 11/80 (13%)
Query 66 NTSICATCQFLRGSIAGTIAKTVVYPLDRLKMKLQV-----QSASVS----SSFSLRH-- 114
N + + G+IAGT+ +T+ YPLD ++ ++Q+ SA V+ S+ SL +
Sbjct 239 NNELTVVTRLTCGAIAGTVGQTIAYPLDVIRRRMQMVGWKDASAIVTGEGRSTASLEYTG 298
Query 115 VPRELQTFVQTGGFFSLWKG 134
+ + V+ GF +L+KG
Sbjct 299 MVDAFRKTVRHEGFGALYKG 318
> YPR011c
Length=326
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/48 (43%), Positives = 32/48 (66%), Gaps = 4/48 (8%)
Query 75 FLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSS----FSLRHVPRE 118
FL G +AG +++TVV P +R+K+ LQVQS++ S + S+R V E
Sbjct 26 FLAGGVAGAVSRTVVSPFERVKILLQVQSSTTSYNRGIFSSIRQVYHE 73
Score = 30.8 bits (68), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 11/24 (45%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 78 GSIAGTIAKTVVYPLDRLKMKLQV 101
G+I+G +A+T+ YP D L+ + QV
Sbjct 240 GAISGGVAQTITYPFDLLRRRFQV 263
> Hs17445255
Length=213
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 34/64 (53%), Gaps = 2/64 (3%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSAS--VSSSFSLRHVPRELQTFVQTGGFFSL 131
FL G +A I+K V P+ R+K+ LQVQ AS V++ + + + + G S
Sbjct 11 DFLAGGVAAAISKMAVVPIQRVKLLLQVQHASKQVTADKQYKGIIDCVVCISKEQGVLSF 70
Query 132 WKGN 135
W+GN
Sbjct 71 WRGN 74
> At3g53940
Length=358
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/66 (37%), Positives = 35/66 (53%), Gaps = 7/66 (10%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQ----SASVSSSFSLRHVPRELQTFVQTGGFF 129
+ L G IAG +KT PL RL + Q+Q A++ SS ++ H E V+ GF
Sbjct 72 RLLAGGIAGAFSKTCTAPLARLTILFQIQGMQSEAAILSSPNIWH---EASRIVKEEGFR 128
Query 130 SLWKGN 135
+ WKGN
Sbjct 129 AFWKGN 134
Score = 32.3 bits (72), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 18/69 (26%), Positives = 29/69 (42%), Gaps = 11/69 (15%)
Query 66 NTSICATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQT 125
N + + F+ G +AG A + YPLD ++ +L Q V +T +
Sbjct 171 NAGVDISVHFVSGGLAGLTAASATYPLDLVRTRLSAQG-----------VGHAFRTICRE 219
Query 126 GGFFSLWKG 134
G L+KG
Sbjct 220 EGILGLYKG 228
Score = 28.5 bits (62), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 9/27 (33%), Positives = 20/27 (74%), Gaps = 0/27 (0%)
Query 78 GSIAGTIAKTVVYPLDRLKMKLQVQSA 104
GS++G ++ T +PLD ++ ++Q++ A
Sbjct 271 GSLSGIVSSTATFPLDLVRRRMQLEGA 297
> At2g37890
Length=348
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 32/63 (50%), Gaps = 1/63 (1%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSA-SVSSSFSLRHVPRELQTFVQTGGFFSLW 132
L G IAG I+KT PL RL + Q+Q S + S ++ RE + G+ + W
Sbjct 44 NLLAGGIAGAISKTCTAPLARLTILFQLQGMQSEGAVLSRPNLRREASRIINEEGYRAFW 103
Query 133 KGN 135
KGN
Sbjct 104 KGN 106
Score = 36.2 bits (82), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 32/69 (46%), Gaps = 4/69 (5%)
Query 66 NTSICATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQT 125
NTS F+ G +AG A T YPLD ++ +L Q ++ + + +T +
Sbjct 143 NTSGNPIVHFVSGGLAGITAATATYPLDLVRTRLAAQRNAI----YYQGIEHTFRTICRE 198
Query 126 GGFFSLWKG 134
G L+KG
Sbjct 199 EGILGLYKG 207
> Hs13259541
Length=309
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 36/69 (52%), Gaps = 5/69 (7%)
Query 71 ATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSAS-----VSSSFSLRHVPRELQTFVQT 125
AT +FL A IA + +PLD K++LQ+Q S ++S R V + T V+T
Sbjct 13 ATVKFLGAGTAACIADLITFPLDTAKVRLQIQGESQGPVRATASAQYRGVMGTILTMVRT 72
Query 126 GGFFSLWKG 134
G SL+ G
Sbjct 73 EGPRSLYNG 81
Score = 32.3 bits (72), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 28/63 (44%), Gaps = 1/63 (1%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSLWK 133
+ L GS G +A V P D +K++ Q Q A + +T + GF LWK
Sbjct 119 RLLAGSTTGALAVAVAQPTDVVKVRFQAQ-ARAGGGRRYQSTVNAYKTIAREEGFRGLWK 177
Query 134 GNS 136
G S
Sbjct 178 GTS 180
> YGR096w
Length=314
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/68 (27%), Positives = 38/68 (55%), Gaps = 1/68 (1%)
Query 68 SICATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGG 127
++ A L G+++G +A+++ P+D +K++LQ+ A+ F V ++ ++ G
Sbjct 13 NVAAWKTLLAGAVSGLLARSITAPMDTIKIRLQLTPANGLKPFG-SQVMEVARSMIKNEG 71
Query 128 FFSLWKGN 135
S WKGN
Sbjct 72 IRSFWKGN 79
Score = 30.8 bits (68), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 25/45 (55%), Gaps = 1/45 (2%)
Query 70 CATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRH 114
AT G+I G IAK + +PL+ ++ ++Q ++ FS RH
Sbjct 211 LATLNHSAGTIGGVIAKIITFPLETIRRRMQFMNSKHLEKFS-RH 254
> HsM4507007
Length=678
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 35/63 (55%), Gaps = 2/63 (3%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQ--SASVSSSFSLRHVPRELQTFVQTGGFFSL 131
+F GS+AG + T VYP+D +K ++Q Q S SV ++ + ++ GFF L
Sbjct 329 RFTLGSVAGAVGATAVYPIDLVKTRMQNQRGSGSVVGELMYKNSFDCFKKVLRYEGFFGL 388
Query 132 WKG 134
++G
Sbjct 389 YRG 391
Score = 28.5 bits (62), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 35/126 (27%), Positives = 53/126 (42%), Gaps = 21/126 (16%)
Query 13 CRPGVERSEGLFGLF-GFFGE--DEKNRKRIKMTADEFRVSPQAAAAGGVEVSRHLNTSI 69
C V R EG FGL+ G + K IK+T ++F + +R + S+
Sbjct 375 CFKKVLRYEGFFGLYRGLIPQLIGVAPEKAIKLTVNDFVRD---------KFTRR-DGSV 424
Query 70 CATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQ-SASVSSSFSLRHVPRELQTFVQTGGF 128
+ L G AG PL+ +K++LQV + S +V R+L G
Sbjct 425 PLPAEVLAGGCAGGSQVIFTNPLEIVKIRLQVAGEITTGPRVSALNVLRDL-------GI 477
Query 129 FSLWKG 134
F L+KG
Sbjct 478 FGLYKG 483
Score = 28.5 bits (62), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 33/64 (51%), Gaps = 12/64 (18%)
Query 78 GSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFS-----LRHVPRELQTFVQTGGFFSLW 132
G++AG A ++V P D +K +LQV + + +++S R + RE G + W
Sbjct 525 GAMAGVPAASLVTPADVIKTRLQVAARAGQTTYSGVIDCFRKILRE-------EGPSAFW 577
Query 133 KGNS 136
KG +
Sbjct 578 KGTA 581
> Hs21361103
Length=678
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 35/63 (55%), Gaps = 2/63 (3%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQ--SASVSSSFSLRHVPRELQTFVQTGGFFSL 131
+F GS+AG + T VYP+D +K ++Q Q S SV ++ + ++ GFF L
Sbjct 329 RFTLGSVAGAVGATAVYPIDLVKTRMQNQRGSGSVVGELMYKNSFDCFKKVLRYEGFFGL 388
Query 132 WKG 134
++G
Sbjct 389 YRG 391
Score = 28.5 bits (62), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 35/126 (27%), Positives = 53/126 (42%), Gaps = 21/126 (16%)
Query 13 CRPGVERSEGLFGLF-GFFGE--DEKNRKRIKMTADEFRVSPQAAAAGGVEVSRHLNTSI 69
C V R EG FGL+ G + K IK+T ++F + +R + S+
Sbjct 375 CFKKVLRYEGFFGLYRGLIPQLIGVAPEKAIKLTVNDFVRD---------KFTRR-DGSV 424
Query 70 CATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQ-SASVSSSFSLRHVPRELQTFVQTGGF 128
+ L G AG PL+ +K++LQV + S +V R+L G
Sbjct 425 PLPAEVLAGGCAGGSQVIFTNPLEIVKIRLQVAGEITTGPRVSALNVLRDL-------GI 477
Query 129 FSLWKG 134
F L+KG
Sbjct 478 FGLYKG 483
Score = 28.5 bits (62), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 33/64 (51%), Gaps = 12/64 (18%)
Query 78 GSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFS-----LRHVPRELQTFVQTGGFFSLW 132
G++AG A ++V P D +K +LQV + + +++S R + RE G + W
Sbjct 525 GAMAGVPAASLVTPADVIKTRLQVAARAGQTTYSGVIDCFRKILRE-------EGPSAFW 577
Query 133 KGNS 136
KG +
Sbjct 578 KGTA 581
> YOR222w
Length=307
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 40/70 (57%), Gaps = 6/70 (8%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRE------LQTFVQTGG 127
QF+ G++AG TV+YPLD +K + Q++ + +++ + V R L+ V+ G
Sbjct 15 QFISGAVAGISELTVMYPLDVVKTRFQLEVTTPTAAAVGKQVERYNGVIDCLKKIVKKEG 74
Query 128 FFSLWKGNST 137
F L++G S+
Sbjct 75 FSRLYRGISS 84
> Hs20541861
Length=154
Score = 38.5 bits (88), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/72 (27%), Positives = 38/72 (52%), Gaps = 2/72 (2%)
Query 66 NTSICATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSAS--VSSSFSLRHVPRELQTFV 123
+ ++ T FL G +A I+K V P++R+K+ L+VQ AS +++ + + + +
Sbjct 3 DAAVSFTKDFLAGGVAAAISKMGVAPIERVKLLLEVQHASKQITADMQYKGIIDCVVHIL 62
Query 124 QTGGFFSLWKGN 135
+ G S W N
Sbjct 63 KEQGVLSFWHRN 74
> CE14263
Length=300
Score = 38.1 bits (87), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 35/67 (52%), Gaps = 16/67 (23%)
Query 78 GSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRH---------VPRELQTFVQTGGF 128
G A ++KT V P++R+K+ LQVQ AS + + R+ VP+E G
Sbjct 19 GGTAAAVSKTAVAPIERVKLLLQVQDASKAIAVDKRYKGIMDVLIRVPKE-------QGV 71
Query 129 FSLWKGN 135
+LW+GN
Sbjct 72 AALWRGN 78
> 7301942
Length=695
Score = 38.1 bits (87), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 36/65 (55%), Gaps = 1/65 (1%)
Query 71 ATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSA-SVSSSFSLRHVPRELQTFVQTGGFF 129
++ +F GS AG + TVVYP+D +K ++Q Q A S + R+ + V+ GF
Sbjct 342 SSYRFTLGSFAGAVGATVVYPIDLVKTRMQNQRAGSYIGEVAYRNSWDCFKKVVRHEGFM 401
Query 130 SLWKG 134
L++G
Sbjct 402 GLYRG 406
Score = 28.5 bits (62), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 32/59 (54%), Gaps = 2/59 (3%)
Query 78 GSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSLWKGNS 136
G+IAG A ++V P D +K +LQV + S ++++ V + + G + WKG +
Sbjct 540 GAIAGVPAASLVTPADVIKTRLQVVARSGQTTYT--GVWDATKKIMAEEGPRAFWKGTA 596
Score = 27.7 bits (60), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 35/127 (27%), Positives = 51/127 (40%), Gaps = 23/127 (18%)
Query 13 CRPGVERSEGLFGLF-GFFGE--DEKNRKRIKMTADEFRVSPQAAAAGGVEVSRHLNTSI 69
C V R EG GL+ G + K IK+T ++ G +I
Sbjct 390 CFKKVVRHEGFMGLYRGLLPQLMGVAPEKAIKLTVNDLVRDKLTDKKG----------NI 439
Query 70 CATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLR--HVPRELQTFVQTGG 127
+ L G AG PL+ +K++LQV + ++S +R V REL G
Sbjct 440 PTWAEVLAGGCAGASQVVFTNPLEIVKIRLQV-AGEIASGSKIRAWSVVREL-------G 491
Query 128 FFSLWKG 134
F L+KG
Sbjct 492 LFGLYKG 498
> At5g51050
Length=487
Score = 38.1 bits (87), Expect = 0.005, Method: Composition-based stats.
Identities = 22/75 (29%), Positives = 36/75 (48%), Gaps = 11/75 (14%)
Query 61 VSRHLNTSICATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQ 120
+S+H+ S F+ G IAG ++T PLDRLK+ LQ+Q + ++
Sbjct 202 ISKHIKRS----NYFIAGGIAGAASRTATAPLDRLKVLLQIQKTDA-------RIREAIK 250
Query 121 TFVQTGGFFSLWKGN 135
+ GG ++GN
Sbjct 251 LIWKQGGVRGFFRGN 265
Score = 33.5 bits (75), Expect = 0.12, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 29/57 (50%), Gaps = 7/57 (12%)
Query 78 GSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSLWKG 134
G+I+G + T VYPL ++ ++Q + A S S R + G+ +L+KG
Sbjct 409 GTISGALGATCVYPLQVVRTRMQAERARTSMSGVFRRT-------ISEEGYRALYKG 458
Score = 33.5 bits (75), Expect = 0.14, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 26/49 (53%), Gaps = 5/49 (10%)
Query 69 ICATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPR 117
I T + G +AG +A+ +YPLD +K +LQ ++ + VPR
Sbjct 301 IGTTVRLFAGGMAGAVAQASIYPLDLVKTRLQTYTSQAGVA-----VPR 344
> At3g20240
Length=348
Score = 38.1 bits (87), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 32/62 (51%), Gaps = 7/62 (11%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSLWK 133
+FL G++AG + K V+ PL+ ++ ++ V S R +P VQ G+ LW
Sbjct 51 EFLSGALAGAMTKAVLAPLETIRTRMIVGVGS-------RSIPGSFLEVVQKQGWQGLWA 103
Query 134 GN 135
GN
Sbjct 104 GN 105
> CE05946
Length=588
Score = 38.1 bits (87), Expect = 0.006, Method: Composition-based stats.
Identities = 22/67 (32%), Positives = 30/67 (44%), Gaps = 14/67 (20%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSAS-----VSSSFSLRHVPRELQTFVQTGGF 128
+ G AG +++T P DR+K+ LQV S+ V S L H GG
Sbjct 251 HLVAGGAAGAVSRTCTAPFDRIKVYLQVNSSKTNRLGVMSCLKLLHA---------EGGI 301
Query 129 FSLWKGN 135
S W+GN
Sbjct 302 KSFWRGN 308
> YPR021c
Length=902
Score = 37.4 bits (85), Expect = 0.009, Method: Composition-based stats.
Identities = 16/30 (53%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 75 FLRGSIAGTIAKTVVYPLDRLKMKLQVQSA 104
F GSIAG I TVVYP+D +K ++Q Q +
Sbjct 534 FSLGSIAGCIGATVVYPIDFIKTRMQAQRS 563
> SPAC17H9.08
Length=326
Score = 37.4 bits (85), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 33/62 (53%), Gaps = 1/62 (1%)
Query 76 LRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRH-VPRELQTFVQTGGFFSLWKG 134
+ G AG +AK+VV PLDR+K+ Q AS RH + + ++ G L++G
Sbjct 22 IAGGTAGCVAKSVVAPLDRVKILYQTNHASYRGYAYSRHGLYKAIKHIYHVYGLHGLYQG 81
Query 135 NS 136
++
Sbjct 82 HT 83
Score = 27.3 bits (59), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 11/26 (42%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKL 99
+FL GS+AGT + YPL+ ++++L
Sbjct 119 RFLSGSLAGTCSVFFTYPLELIRVRL 144
> CE29222
Length=313
Score = 37.0 bits (84), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 14/25 (56%), Positives = 22/25 (88%), Gaps = 0/25 (0%)
Query 78 GSIAGTIAKTVVYPLDRLKMKLQVQ 102
G++AGT+AKTV+YPLD ++ +LQ+
Sbjct 219 GAMAGTVAKTVLYPLDMVRHRLQMN 243
Score = 33.9 bits (76), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 18/67 (26%), Positives = 31/67 (46%), Gaps = 0/67 (0%)
Query 66 NTSICATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQT 125
N S+ +T F G+++G +A T PLD ++ +L Q A + H + +
Sbjct 114 NQSVRSTSDFACGALSGCLAMTAAMPLDVIRTRLVAQKAGHAVYTGTMHAVKHIWEKEGI 173
Query 126 GGFFSLW 132
G+F W
Sbjct 174 AGYFRGW 180
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 15/60 (25%), Positives = 30/60 (50%), Gaps = 2/60 (3%)
Query 78 GSIAGTIAKTVVYPLDRLKMKLQVQSASV--SSSFSLRHVPRELQTFVQTGGFFSLWKGN 135
G +G + + ++ PLD LK++ Q+Q + S + V + + + G + WKG+
Sbjct 22 GLASGIVTRMIIQPLDVLKIRFQLQEEPIRGKKSGKYKGVMQSIFLITREEGAHAFWKGH 81
> At3g55640
Length=332
Score = 37.0 bits (84), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 35/71 (49%), Gaps = 3/71 (4%)
Query 67 TSICATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRH--VPRELQTFVQ 124
+ I + Q L G +AG +KT PL RL + QVQ +++ +LR + E +
Sbjct 30 SHIESASQLLAGGLAGAFSKTCTAPLSRLTILFQVQGMHTNAA-ALRKPSILHEASRILN 88
Query 125 TGGFFSLWKGN 135
G + WKGN
Sbjct 89 EEGLKAFWKGN 99
Score = 33.5 bits (75), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 31/65 (47%), Gaps = 4/65 (6%)
Query 73 CQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSLW 132
F+ G +AG A + YPLD ++ +L Q+ + S + H R + T G L+
Sbjct 143 VHFVAGGLAGITAASATYPLDLVRTRLAAQTKVIYYS-GIWHTLRSITT---DEGILGLY 198
Query 133 KGNST 137
KG T
Sbjct 199 KGLGT 203
> Hs21687151
Length=341
Score = 37.0 bits (84), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 32/62 (51%), Gaps = 3/62 (4%)
Query 75 FLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSLWKG 134
+ GS+AG ++ V YP D +K +L +Q+ S L H T Q GF +L++G
Sbjct 110 IMAGSLAGMVSTIVTYPTDLIKTRLIMQNILEPSYRGLLH---AFSTIYQQEGFLALYRG 166
Query 135 NS 136
S
Sbjct 167 VS 168
> Hs20548100
Length=341
Score = 37.0 bits (84), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 32/62 (51%), Gaps = 3/62 (4%)
Query 75 FLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSLWKG 134
+ GS+AG ++ V YP D +K +L +Q+ S L H T Q GF +L++G
Sbjct 110 IMAGSLAGMVSTIVTYPTDLIKTRLIMQNILEPSYRGLLH---AFSTIYQQEGFLALYRG 166
Query 135 NS 136
S
Sbjct 167 VS 168
> 7292557
Length=299
Score = 37.0 bits (84), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 35/71 (49%), Gaps = 16/71 (22%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRH---------VPRELQTFVQ 124
F G I+ ++KT V P++R+K+ LQVQ S S ++ +P+E
Sbjct 13 DFAAGGISAAVSKTAVAPIERVKLLLQVQHISKQISPDKQYKGMVDCFIRIPKEQ----- 67
Query 125 TGGFFSLWKGN 135
GF S W+GN
Sbjct 68 --GFSSFWRGN 76
Score = 27.3 bits (59), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 28/126 (22%), Positives = 53/126 (42%), Gaps = 17/126 (13%)
Query 10 LRRCRPGVERSEGLFGLFGFFGEDEKNRKRIKMTADEFRVSPQAAAAGGVEVSRHLNTSI 69
L C + +S+G+ GL+ FG + + +AA G + +R +
Sbjct 158 LGNCLTKIFKSDGIVGLYRGFGVSVQGI-----------IIYRAAYFGFYDTARGMLPDP 206
Query 70 CATCQFLRGSIA---GTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQ-- 124
T ++ +IA T+A V YP D ++ ++ +QS ++ ++ T +
Sbjct 207 KNTPIYISWAIAQVVTTVAGIVSYPFDTVRRRMMMQSGRKATEVIYKNTLHCWATIAKQE 266
Query 125 -TGGFF 129
TG FF
Sbjct 267 GTGAFF 272
> Hs7706150
Length=347
Score = 36.6 bits (83), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 21/79 (26%), Positives = 41/79 (51%), Gaps = 2/79 (2%)
Query 56 AGGVEVSRHLNTSICATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHV 115
A G E +L TS + G++AG + +V+YP+D +K ++Q S + ++ +
Sbjct 30 ATGSEDYENLPTSASVSTHMTAGAMAGILEHSVMYPVDSVKTRMQSLSPDPKAQYT--SI 87
Query 116 PRELQTFVQTGGFFSLWKG 134
L+ ++T GF+ +G
Sbjct 88 YGALKKIMRTEGFWRPLRG 106
> YBL030c
Length=318
Score = 36.6 bits (83), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 32/66 (48%), Gaps = 3/66 (4%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTG---GFFS 130
FL G ++ +AKT P++R+K+ +Q Q + R L F +T G S
Sbjct 26 DFLMGGVSAAVAKTAASPIERVKLLIQNQDEMLKQGTLDRKYAGILDCFKRTATQEGVIS 85
Query 131 LWKGNS 136
W+GN+
Sbjct 86 FWRGNT 91
> SPAC8C9.12c
Length=303
Score = 36.6 bits (83), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 37/64 (57%), Gaps = 2/64 (3%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSLWK 133
L G+ +G + +V+YP+D +K ++Q+ + VS S S ++ + T G +SLW+
Sbjct 22 HLLAGAFSGILEHSVMYPVDAIKTRMQMLNG-VSRSVS-GNIVNSVIKISSTEGVYSLWR 79
Query 134 GNST 137
G S+
Sbjct 80 GISS 83
> Hs17436303
Length=419
Score = 35.8 bits (81), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 32/58 (55%), Gaps = 4/58 (6%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSL 131
+ + IA +A+T PLDRLK+ +QV S S +R + L+ V+ GG FSL
Sbjct 185 RLVSAGIASAVARTCTAPLDRLKVMMQVHSL---KSRKMRLI-SGLEQLVKEGGIFSL 238
> YBR085w
Length=307
Score = 35.8 bits (81), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 32/66 (48%), Gaps = 3/66 (4%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTG---GFFS 130
FL G ++ IAKT P++R+K+ +Q Q + + + F +T G S
Sbjct 15 NFLMGGVSAAIAKTAASPIERVKILIQNQDEMIKQGTLDKKYSGIVDCFKRTAKQEGLIS 74
Query 131 LWKGNS 136
W+GN+
Sbjct 75 FWRGNT 80
> Hs22046149
Length=469
Score = 35.4 bits (80), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 34/62 (54%), Gaps = 4/62 (6%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSLWK 133
+ G AG +++T PLDRLK+ +QV AS S++ + ++ GG SLW+
Sbjct 189 HLVAGGGAGAVSRTCTAPLDRLKVLMQVH-ASRSNNMG---IVGGFTQMIREGGARSLWR 244
Query 134 GN 135
GN
Sbjct 245 GN 246
Score = 28.5 bits (62), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 10/26 (38%), Positives = 20/26 (76%), Gaps = 0/26 (0%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKL 99
+ + GS+AG IA++ +YP++ LK ++
Sbjct 283 RLVAGSLAGAIAQSSIYPMEVLKTRM 308
> CE02298
Length=298
Score = 35.4 bits (80), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 29/60 (48%), Gaps = 0/60 (0%)
Query 76 LRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFSLRHVPRELQTFVQTGGFFSLWKGN 135
L GS A I+KT P DR+K+ LQ+Q S + + + G +LW+GN
Sbjct 18 LAGSAAAAISKTTTAPFDRVKLVLQLQRQSEFAMAEYNGIRDCISKIRLEQGAMALWRGN 77
> 7291871
Length=323
Score = 35.4 bits (80), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 22/29 (75%), Gaps = 0/29 (0%)
Query 75 FLRGSIAGTIAKTVVYPLDRLKMKLQVQS 103
FL G+++G +AK +VYP D LK ++Q+ +
Sbjct 216 FLNGALSGVLAKMIVYPADLLKKRIQLMA 244
> Hs7657581
Length=675
Score = 35.0 bits (79), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 34/63 (53%), Gaps = 2/63 (3%)
Query 74 QFLRGSIAGTIAKTVVYPLDRLKMKLQVQ--SASVSSSFSLRHVPRELQTFVQTGGFFSL 131
+F GS+AG + T VYP+D +K ++Q Q + S ++ + ++ GFF L
Sbjct 331 RFGLGSVAGAVGATAVYPIDLVKTRMQNQRSTGSFVGELMYKNSFDCFKKVLRYEGFFGL 390
Query 132 WKG 134
++G
Sbjct 391 YRG 393
Score = 32.7 bits (73), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 35/126 (27%), Positives = 52/126 (41%), Gaps = 21/126 (16%)
Query 13 CRPGVERSEGLFGLF-GFFGE--DEKNRKRIKMTADEFRVSPQAAAAGGVEVSRHLNTSI 69
C V R EG FGL+ G + K IK+T ++F + H + S+
Sbjct 377 CFKKVLRYEGFFGLYRGLLPQLLGVAPEKAIKLTVNDFVR----------DKFMHKDGSV 426
Query 70 CATCQFLRGSIAGTIAKTVVYPLDRLKMKLQVQ-SASVSSSFSLRHVPRELQTFVQTGGF 128
+ L G AG PL+ +K++LQV + S V R+L GF
Sbjct 427 PLAAEILAGGCAGGSQVIFTNPLEIVKIRLQVAGEITTGPRVSALSVVRDL-------GF 479
Query 129 FSLWKG 134
F ++KG
Sbjct 480 FGIYKG 485
Score = 32.0 bits (71), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 34/65 (52%), Gaps = 12/65 (18%)
Query 75 FLRGSIAGTIAKTVVYPLDRLKMKLQVQSASVSSSFS-----LRHVPRELQTFVQTGGFF 129
L G+IAG A ++V P D +K +LQV + + +++S R + RE G
Sbjct 524 LLAGAIAGMPAASLVTPADVIKTRLQVAARAGQTTYSGVIDCFRKILRE-------EGPK 576
Query 130 SLWKG 134
+LWKG
Sbjct 577 ALWKG 581
> CE14428
Length=300
Score = 35.0 bits (79), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query 78 GSIAGTIAKTVVYPLDRLKMKLQVQSAS--VSSSFSLRHVPRELQTFVQTGGFFSLWKGN 135
G A I+KT V P++R+K+ LQV S V++ + + L + G+ + W+GN
Sbjct 19 GGTAAAISKTAVAPIERVKLLLQVSDVSETVTADKKYKGIMDVLARVPKEQGYAAFWRGN 78
Lambda K H
0.322 0.135 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1461889840
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40