bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3074_orf1
Length=159
Score E
Sequences producing significant alignments: (Bits) Value
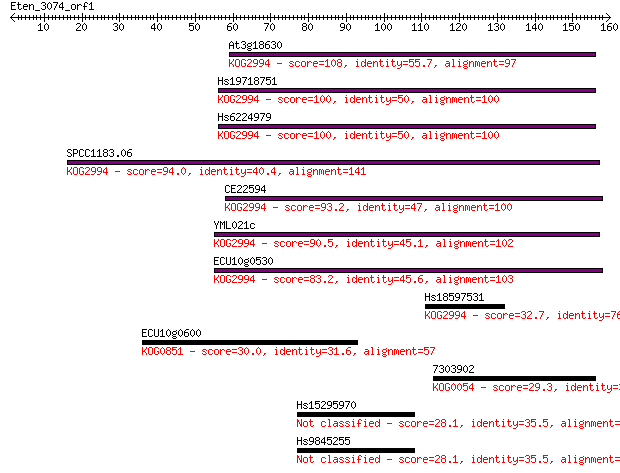
At3g18630 108 5e-24
Hs19718751 100 1e-21
Hs6224979 100 2e-21
SPCC1183.06 94.0 1e-19
CE22594 93.2 2e-19
YML021c 90.5 1e-18
ECU10g0530 83.2 2e-16
Hs18597531 32.7 0.34
ECU10g0600 30.0 2.2
7303902 29.3 3.1
Hs15295970 28.1 7.5
Hs9845255 28.1 8.2
> At3g18630
Length=330
Score = 108 bits (270), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 54/101 (53%), Positives = 67/101 (66%), Gaps = 6/101 (5%)
Query 59 DSWYEALQGELQKPYFRKCLLAVKEQREKFPSS----IYPPQELMFKAFRSTPLDKVKVV 114
+SW +AL GE KPY + L+ +RE S IYPPQ L+F A +TP D+VK V
Sbjct 111 ESWLKALPGEFHKPYAKS--LSDFLEREIITDSKSPLIYPPQHLIFNALNTTPFDRVKTV 168
Query 115 IVGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDRSI 155
I+GQDPYH PGQAMGL FSVP G + P SL NIFKE+ + +
Sbjct 169 IIGQDPYHGPGQAMGLSFSVPEGEKLPSSLLNIFKELHKDV 209
> Hs19718751
Length=313
Score = 100 bits (250), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 50/100 (50%), Positives = 64/100 (64%), Gaps = 2/100 (2%)
Query 56 AMGDSWYEALQGELQKPYFRKCLLAVKEQREKFPSSIYPPQELMFKAFRSTPLDKVKVVI 115
G+SW + L GE KPYF K + V E+R+ + ++YPP +F + + VKVVI
Sbjct 93 GFGESWKKHLSGEFGKPYFIKLMGFVAEERKHY--TVYPPPHQVFTWTQMCDIKDVKVVI 150
Query 116 VGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDRSI 155
+GQDPYH P QA GLCFSV R V PPPSL NI+KE+ I
Sbjct 151 LGQDPYHGPNQAHGLCFSVQRPVPPPPSLENIYKELSTDI 190
> Hs6224979
Length=304
Score = 100 bits (248), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 50/100 (50%), Positives = 64/100 (64%), Gaps = 2/100 (2%)
Query 56 AMGDSWYEALQGELQKPYFRKCLLAVKEQREKFPSSIYPPQELMFKAFRSTPLDKVKVVI 115
G+SW + L GE KPYF K + V E+R+ + ++YPP +F + + VKVVI
Sbjct 84 GFGESWKKHLSGEFGKPYFIKLMGFVAEERKHY--TVYPPPHQVFTWTQMCDIKDVKVVI 141
Query 116 VGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDRSI 155
+GQDPYH P QA GLCFSV R V PPPSL NI+KE+ I
Sbjct 142 LGQDPYHGPNQAHGLCFSVQRPVPPPPSLENIYKELSTDI 181
> SPCC1183.06
Length=322
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 57/150 (38%), Positives = 80/150 (53%), Gaps = 13/150 (8%)
Query 16 SRSSSPAAAAATGAAGAAASAVSPFGTFV----STAAAEKMWR---EAMGDSWYEALQGE 68
+ +SSPA ++VS F T A K+ + + + SW++AL+ E
Sbjct 34 TNTSSPALKDTQVLDNKENNSVSKFNKEKWAENLTPAQRKLLQLEIDTLESSWFDALKDE 93
Query 69 LQKPYFRKCLLAVKE--QREKFPSSIYPPQELMFKAFRSTPLDKVKVVIVGQDPYHQPGQ 126
KPYF L +KE +E ++PP+E ++ TPL K KV+++GQDPYH GQ
Sbjct 94 FLKPYF----LNLKEFLMKEWQSQRVFPPKEDIYSWSHHTPLHKTKVILLGQDPYHNIGQ 149
Query 127 AMGLCFSVPRGVRPPPSLTNIFKEIDRSIP 156
A GLCFSV G+ PPSL NI+K I P
Sbjct 150 AHGLCFSVRPGIPCPPSLVNIYKAIKIDYP 179
> CE22594
Length=282
Score = 93.2 bits (230), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 47/107 (43%), Positives = 63/107 (58%), Gaps = 15/107 (14%)
Query 58 GDSWYEALQGELQKPYFRKCLLAVKEQREKFPSS-------IYPPQELMFKAFRSTPLDK 110
G+SW + L+ E +K Y K EKF +S ++PP +F F P D+
Sbjct 63 GESWSKLLEEEFKKGYISKI--------EKFLNSEVNKGKQVFPPPTQIFTTFNLLPFDE 114
Query 111 VKVVIVGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDRSIPG 157
+ VVI+GQDPYH QA GL FSV +GV+PPPSL NI+KE++ I G
Sbjct 115 ISVVIIGQDPYHDDNQAHGLSFSVQKGVKPPPSLKNIYKELESDIEG 161
> YML021c
Length=359
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 46/102 (45%), Positives = 62/102 (60%), Gaps = 2/102 (1%)
Query 55 EAMGDSWYEALQGELQKPYFRKCLLAVKEQREKFPSSIYPPQELMFKAFRSTPLDKVKVV 114
E + DSW+ L E +KPYF K V +E+ +++PP + ++ R TP +KVKVV
Sbjct 100 ETIDDSWFPHLMDEFKKPYFVKLKQFV--TKEQADHTVFPPAKDIYSWTRLTPFNKVKVV 157
Query 115 IVGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDRSIP 156
I+GQDPYH QA GL FSV PPSL NI+KE+ + P
Sbjct 158 IIGQDPYHNFNQAHGLAFSVKPPTPAPPSLKNIYKELKQEYP 199
> ECU10g0530
Length=286
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 47/103 (45%), Positives = 56/103 (54%), Gaps = 5/103 (4%)
Query 55 EAMGDSWYEALQGELQKPYFRKCLLAVKEQREKFPSSIYPPQELMFKAFRSTPLDKVKVV 114
E + W L+ E K YFR + E R P P + +F R PL+K KVV
Sbjct 70 EKIDPRWRMHLKPEFGKDYFRGVKRFLHENRNHLP-----PIDKIFTFSRFFPLEKTKVV 124
Query 115 IVGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDRSIPG 157
I+GQDPYH QAMGL FSVP G+ PPSL NI+ EI I G
Sbjct 125 IMGQDPYHNDNQAMGLSFSVPVGIPVPPSLKNIYLEISSDIEG 167
> Hs18597531
Length=113
Score = 32.7 bits (73), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 16/28 (57%), Positives = 19/28 (67%), Gaps = 7/28 (25%)
Query 111 VKVVIVGQDPYHQPGQAM-------GLC 131
V+VVI+GQDPYH+P QA GLC
Sbjct 28 VEVVILGQDPYHRPYQAHEAALVSKGLC 55
> ECU10g0600
Length=623
Score = 30.0 bits (66), Expect = 2.2, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 30/57 (52%), Gaps = 7/57 (12%)
Query 36 AVSPFGTFVSTAAAEKMWREAMGDSWYEALQGELQKPYFRKCLLAVKEQREKFPSSI 92
A S FG +S + M EA G+ LQ +++ YFR+CL +K +++ + I
Sbjct 546 ATSFFG--ISAREMKVMSEEAPGE-----LQALIRRMYFRECLFRIKSKQDSYNDEI 595
> 7303902
Length=1344
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 17/43 (39%), Positives = 21/43 (48%), Gaps = 1/43 (2%)
Query 113 VVIVGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDRSI 155
V I Q P+ P QAMGL V GVR L N ++R +
Sbjct 1018 VTITCQSPFGYP-QAMGLIDMVQWGVRQTAELENTMTAVERVV 1059
> Hs15295970
Length=474
Score = 28.1 bits (61), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 11/31 (35%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 77 CLLAVKEQREKFPSSIYPPQELMFKAFRSTP 107
C+ A+K Q ++FP+ + P + + +AFR P
Sbjct 122 CMAALKHQPQEFPTYVEPTNDEICEAFRKDP 152
> Hs9845255
Length=474
Score = 28.1 bits (61), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 11/31 (35%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 77 CLLAVKEQREKFPSSIYPPQELMFKAFRSTP 107
C+ A+K Q ++FP+ + P + + +AFR P
Sbjct 122 CMAALKHQPQEFPTYVEPTNDEICEAFRKDP 152
Lambda K H
0.319 0.134 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2136300674
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40