bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3054_orf1
Length=136
Score E
Sequences producing significant alignments: (Bits) Value
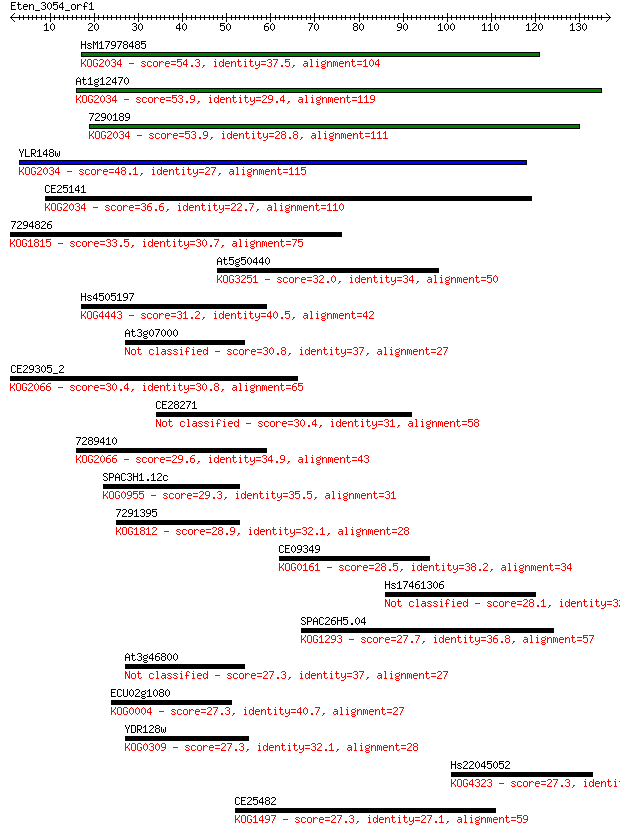
HsM17978485 54.3 7e-08
At1g12470 53.9 8e-08
7290189 53.9 8e-08
YLR148w 48.1 5e-06
CE25141 36.6 0.014
7294826 33.5 0.12
At5g50440 32.0 0.36
Hs4505197 31.2 0.58
At3g07000 30.8 0.82
CE29305_2 30.4 1.1
CE28271 30.4 1.1
7289410 29.6 1.7
SPAC3H1.12c 29.3 2.3
7291395 28.9 3.1
CE09349 28.5 4.3
Hs17461306 28.1 5.2
SPAC26H5.04 27.7 6.9
At3g46800 27.3 8.1
ECU02g1080 27.3 8.7
YDR128w 27.3 9.2
Hs22045052 27.3 9.5
CE25482 27.3 9.6
> HsM17978485
Length=973
Score = 54.3 bits (129), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 39/120 (32%), Positives = 54/120 (45%), Gaps = 16/120 (13%)
Query 17 RCLVIDAGQLCDLCGAAVVCSKFFAFRCGHCMHAACV-QALL--IPAMSTEELGRFDRSV 73
R ++ C C ++ F+ F CGH HA C+ QA+ +PA L R +
Sbjct 843 RYGTVEPQDKCATCDFPLLNRPFYLFLCGHMFHADCLLQAVRPGLPAYKQARLEELQRKL 902
Query 74 AAL------AAAIKEDS-------PEVEALEDALDGLLAQECLLCGSLMIRSLQVPLLGP 120
A +A KE P E L+ LD L+A EC+ CG LMIRS+ P + P
Sbjct 903 GAAPPPAKGSARAKEAEGGAATAGPSREQLKADLDELVAAECVYCGELMIRSIDRPFIDP 962
> At1g12470
Length=994
Score = 53.9 bits (128), Expect = 8e-08, Method: Composition-based stats.
Identities = 35/151 (23%), Positives = 56/151 (37%), Gaps = 32/151 (21%)
Query 16 RRCLVIDAGQLCDLCGAAVV-----------------CSKFFAFRCGHCMHAACVQALLI 58
+R VID + C +C ++ + F+ F CGH HA C+ +
Sbjct 831 QRYAVIDRDEECGVCKRKILMMSGDFRMAQGYSSAGPLAPFYVFPCGHSFHAQCLITHVT 890
Query 59 PAMSTEE---LGRFDRSVAALAAAIKED------------SPEVEALEDALDGLLAQECL 103
E+ + + + L + + D + + L LD +A EC
Sbjct 891 SCAHEEQAEHILDLQKQLTLLGSETRRDINGNRSDEPITSTTTADKLRSELDDAIASECP 950
Query 104 LCGSLMIRSLQVPLLGPADGPLLLDWALRPE 134
CG LMI + +P + P D W LR E
Sbjct 951 FCGELMINEITLPFIKPEDSQYSTSWDLRSE 981
> 7290189
Length=1002
Score = 53.9 bits (128), Expect = 8e-08, Method: Composition-based stats.
Identities = 32/127 (25%), Positives = 61/127 (48%), Gaps = 25/127 (19%)
Query 19 LVIDAGQLCDLCGAAVVCSKFFAFRCGHCMHAACVQALLIPAMSTEELGRFDRSVAALAA 78
L +++ C++C ++ FF F CGH H+ C++ ++P ++ E+ R L A
Sbjct 877 LTVESQDTCEICEMMLLVKPFFIFICGHKFHSDCLEKHVVPLLTKEQCRRLGTLKQQLEA 936
Query 79 AI------------KEDSPEVE----ALEDALDGLLAQECLLCGSLMIRSLQVPLLGPAD 122
+ K+ + E++ AL+ ++ +LA +CL CG L+I ++ D
Sbjct 937 EVQTQAQPQSGALSKQQAMELQRKRAALKTEIEDILAADCLFCG-LLISTI--------D 987
Query 123 GPLLLDW 129
P + DW
Sbjct 988 QPFVDDW 994
> YLR148w
Length=918
Score = 48.1 bits (113), Expect = 5e-06, Method: Composition-based stats.
Identities = 31/115 (26%), Positives = 55/115 (47%), Gaps = 11/115 (9%)
Query 3 KIEIKAEKSPKAARRCLVIDAGQLCDLCGAAVVCSKFFAFRCGHCMHAACVQALLIPAMS 62
K+EI E S K +++ G+ CD CG + KF F CGHC H C+ ++ ++
Sbjct 803 KVEINTEIS-KFNEIYRILEPGKSCDECGKFLQIKKFIVFPCGHCFHWNCIIRVI---LN 858
Query 63 TEELGRFDRSVAALAAAIKEDSPEVEALEDALDGLLAQECLLCGSLMIRSLQVPL 117
+ + ++ L A K + L D L+ ++ ++C LC + I + P+
Sbjct 859 SNDYNLRQKTENFLKAKSKHN------LND-LENIIVEKCGLCSDININKIDQPI 906
> CE25141
Length=1010
Score = 36.6 bits (83), Expect = 0.014, Method: Composition-based stats.
Identities = 25/125 (20%), Positives = 47/125 (37%), Gaps = 15/125 (12%)
Query 9 EKSPKAARRCLVIDAGQLCDLCGAAVVCSKFFAFRCGHCMHAACVQALLIPAMSTEELGR 68
+K K R V+ +C C + F C H H C++ +I +S EE+ +
Sbjct 872 DKQEKLKNRTTVVKPSDVCSHCARPISGRAFNVHSCRHFFHRECLEIAMISFLSQEEVEK 931
Query 69 F-------DRSVAALAA--------AIKEDSPEVEALEDALDGLLAQECLLCGSLMIRSL 113
+R ++ + A E + + + ++ EC LCG++ I +
Sbjct 932 MKTLIIDEERVLSQMKAEQLAGNQKGFIEKQEKYLKIAAFISNIVGAECPLCGNIAISQI 991
Query 114 QVPLL 118
L
Sbjct 992 DKQFL 996
> 7294826
Length=511
Score = 33.5 bits (75), Expect = 0.12, Method: Composition-based stats.
Identities = 23/75 (30%), Positives = 32/75 (42%), Gaps = 5/75 (6%)
Query 1 LNKIEIKAEKSPKAARRCLVIDAGQLCDLCGAAVVCSKFFAFRCGHCMHAACVQALLIPA 60
LN E K E+ A+ C + QLC +C + C + CGH AAC + L
Sbjct 117 LNPFEKKIERESAASTSCAI---PQLCGICFCS--CDELIGLGCGHNFCAACWKQYLANK 171
Query 61 MSTEELGRFDRSVAA 75
+E L + AA
Sbjct 172 TCSEGLANTIKCPAA 186
> At5g50440
Length=219
Score = 32.0 bits (71), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 29/50 (58%), Gaps = 0/50 (0%)
Query 48 MHAACVQALLIPAMSTEELGRFDRSVAALAAAIKEDSPEVEALEDALDGL 97
++++ + LL E+L RFD LA+++K D EV++L +DGL
Sbjct 12 VYSSAKRILLRARNGIEKLERFDSDPTDLASSVKRDITEVQSLCSNMDGL 61
> Hs4505197
Length=5262
Score = 31.2 bits (69), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 17/43 (39%), Positives = 20/43 (46%), Gaps = 10/43 (23%)
Query 17 RCLVIDA-GQLCDLCGAAVVCSKFFAFRCGHCMHAACVQALLI 58
RC V + G+LCDL FF CGH H AC+ L
Sbjct 228 RCAVCEGPGELCDL---------FFCTSCGHHYHGACLDTALT 261
> At3g07000
Length=574
Score = 30.8 bits (68), Expect = 0.82, Method: Composition-based stats.
Identities = 10/27 (37%), Positives = 15/27 (55%), Gaps = 0/27 (0%)
Query 27 CDLCGAAVVCSKFFAFRCGHCMHAACV 53
C+ CG C+ +F F CG +H C+
Sbjct 147 CNACGMLGDCNPYFCFECGFMLHKDCI 173
> CE29305_2
Length=807
Score = 30.4 bits (67), Expect = 1.1, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 32/74 (43%), Gaps = 9/74 (12%)
Query 1 LNKIEIKAEKSPKAARRCLVIDAGQLCDLCGAAVVCS-----KFFA----FRCGHCMHAA 51
LN + + + AA + ++ C LC ++ S K F+ F+CGH H A
Sbjct 719 LNDLNVLTQGLISAADESVSVNIVSRCSLCAQIIINSNQETTKKFSDIKVFKCGHIFHLA 778
Query 52 CVQALLIPAMSTEE 65
C + + S EE
Sbjct 779 CSTSEMERRQSIEE 792
> CE28271
Length=946
Score = 30.4 bits (67), Expect = 1.1, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 27/58 (46%), Gaps = 11/58 (18%)
Query 34 VVCSKFFAFRCGHCMHAACVQALLIPAMSTEELGRFDRSVAALAAAIKEDSPEVEALE 91
+ CS+ F +RC C+ A +A + A + EEL ++E SP E LE
Sbjct 520 LACSRIFGYRCPVCVDANTPKADDVKAPTPEEL-----------VEVREISPVAENLE 566
> 7289410
Length=841
Score = 29.6 bits (65), Expect = 1.7, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 20/48 (41%), Gaps = 5/48 (10%)
Query 16 RRCLVIDAGQLCDLCGAAVVCSK-----FFAFRCGHCMHAACVQALLI 58
RR + QLC LC V+ + CGH H C+Q L+
Sbjct 779 RRGQQVSYEQLCSLCHRPVLMAGTHLYCIIRLECGHVYHKPCIQGELL 826
> SPAC3H1.12c
Length=1131
Score = 29.3 bits (64), Expect = 2.3, Method: Composition-based stats.
Identities = 11/31 (35%), Positives = 14/31 (45%), Gaps = 0/31 (0%)
Query 22 DAGQLCDLCGAAVVCSKFFAFRCGHCMHAAC 52
D + C LCG F+CG C+H C
Sbjct 810 DRKKCCALCGIVGTEGLLVCFKCGTCVHERC 840
> 7291395
Length=509
Score = 28.9 bits (63), Expect = 3.1, Method: Composition-based stats.
Identities = 9/28 (32%), Positives = 16/28 (57%), Gaps = 0/28 (0%)
Query 25 QLCDLCGAAVVCSKFFAFRCGHCMHAAC 52
Q+C +C ++ + KF++ CGH C
Sbjct 151 QMCPVCASSQLGDKFYSLACGHSFCKDC 178
> CE09349
Length=1963
Score = 28.5 bits (62), Expect = 4.3, Method: Composition-based stats.
Identities = 13/34 (38%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 62 STEELGRFDRSVAALAAAIKEDSPEVEALEDALD 95
+ + LGR DR V L + ED E L+D +D
Sbjct 1845 ANKNLGRADRRVRELQFQVDEDKKNFERLQDLID 1878
> Hs17461306
Length=189
Score = 28.1 bits (61), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 11/34 (32%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 86 EVEALEDALDGLLAQECLLCGSLMIRSLQVPLLG 119
+V L+D + G ++C CG++ +RS +P G
Sbjct 84 DVPRLKDEIAGYKPKQCDNCGTITLRSCDIPASG 117
> SPAC26H5.04
Length=729
Score = 27.7 bits (60), Expect = 6.9, Method: Composition-based stats.
Identities = 21/57 (36%), Positives = 26/57 (45%), Gaps = 3/57 (5%)
Query 67 GRFDRSVAALAAAIKEDSPEVEALEDALDGLLAQECLLCGSLMIRSLQVPLLGPADG 123
G D + L A KE+ E+ L D L A ECLL S RS+ + G AD
Sbjct 372 GFIDAAKYLLEYATKEEENEINFLSDELQSSAACECLLSLS---RSVYILRTGLADA 425
> At3g46800
Length=682
Score = 27.3 bits (59), Expect = 8.1, Method: Composition-based stats.
Identities = 10/27 (37%), Positives = 15/27 (55%), Gaps = 0/27 (0%)
Query 27 CDLCGAAVVCSKFFAFRCGHCMHAACV 53
C+ CG A S +F +C +H AC+
Sbjct 274 CNACGTAGDRSPYFCLQCNFMIHRACI 300
> ECU02g1080
Length=152
Score = 27.3 bits (59), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 11/28 (39%), Positives = 14/28 (50%), Gaps = 1/28 (3%)
Query 24 GQLCDLCGAAVVCSKF-FAFRCGHCMHA 50
G C++CG V C K F+CG C
Sbjct 117 GVSCEVCGTEVFCGKIQSGFKCGRCYRV 144
> YDR128w
Length=1148
Score = 27.3 bits (59), Expect = 9.2, Method: Composition-based stats.
Identities = 9/28 (32%), Positives = 14/28 (50%), Gaps = 0/28 (0%)
Query 27 CDLCGAAVVCSKFFAFRCGHCMHAACVQ 54
C+ C V S F C H +H++C +
Sbjct 1098 CNYCDLRVTRSSFICGNCQHVLHSSCAR 1125
> Hs22045052
Length=580
Score = 27.3 bits (59), Expect = 9.5, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 18/35 (51%), Gaps = 3/35 (8%)
Query 101 ECLLCGSLMI---RSLQVPLLGPADGPLLLDWALR 132
E L+CG + + +P+ G AD PLL W R
Sbjct 112 EILICGKCGLGYHQQCHIPIAGSADQPLLTPWFCR 146
> CE25482
Length=412
Score = 27.3 bits (59), Expect = 9.6, Method: Composition-based stats.
Identities = 16/59 (27%), Positives = 30/59 (50%), Gaps = 1/59 (1%)
Query 52 CVQALLIPAMSTEELGRFDRSVAALAAAIKEDSPEVEALEDALDGLLAQECLLCGSLMI 110
C+ L++ ++ +E GR + AL A + SP+ + A +G AQ C+ L++
Sbjct 111 CILRLMLASLYEKE-GRIKDAAQALIAINSDTSPKFNGPQAAKEGAKAQLCIRITKLLL 168
Lambda K H
0.324 0.137 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1425342594
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40