bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3038_orf2
Length=155
Score E
Sequences producing significant alignments: (Bits) Value
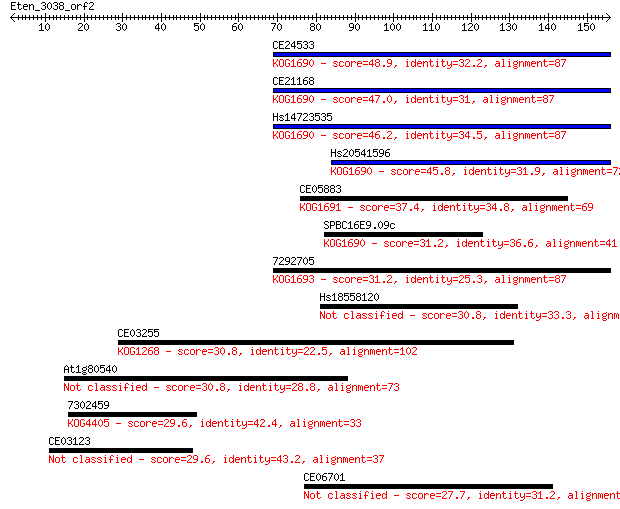
CE24533 48.9 4e-06
CE21168 47.0 1e-05
Hs14723535 46.2 3e-05
Hs20541596 45.8 4e-05
CE05883 37.4 0.012
SPBC16E9.09c 31.2 0.75
7292705 31.2 0.79
Hs18558120 30.8 0.96
CE03255 30.8 1.0
At1g80540 30.8 1.2
7302459 29.6 2.2
CE03123 29.6 2.5
CE06701 27.7 10.0
> CE24533
Length=211
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 47/97 (48%), Gaps = 13/97 (13%)
Query 69 LVLALGYFAHSVSAAYFYVQESQDKCFIESVPTGVALTASYKNH----------ENPGVT 118
L++AL A + YF++ E++ KCFIE +P +T +YK + P +
Sbjct 3 LLIALLSLATYADSLYFHIAETEKKCFIEEIPDETMVTGNYKVQLYDPNTKGYGDYPNIG 62
Query 119 CSIIFKDPSGRSVYSREVLPADAEGKVTHMTTTTGEY 155
+ KDP + + S+ AEG+ T + T GE+
Sbjct 63 MHVEVKDPEDKVILSK---LYTAEGRFTFTSNTPGEH 96
> CE21168
Length=167
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 47/97 (48%), Gaps = 13/97 (13%)
Query 69 LVLALGYFAHSVSAAYFYVQESQDKCFIESVPTGVALTASYKNH----------ENPGVT 118
L++AL A + YF++ E++ KCFIE +P +T +YK + P +
Sbjct 3 LLIALLSLATYADSLYFHIAETEKKCFIEEIPDETMVTGNYKVQLYDPNTKGYGDYPNIG 62
Query 119 CSIIFKDPSGRSVYSREVLPADAEGKVTHMTTTTGEY 155
+ KDP + + S+ +EG+ T + T GE+
Sbjct 63 MHVEVKDPEDKVILSKLYT---SEGRFTFTSNTPGEH 96
> Hs14723535
Length=214
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 30/98 (30%), Positives = 49/98 (50%), Gaps = 15/98 (15%)
Query 69 LVLALGYFAHSVSAAYFYVQESQDKCFIESVPTGVALTASY------KNHEN-----PGV 117
L+L L + A SA YF++ E++ KCFIE +P + +Y K E PG+
Sbjct 4 LLLVL-WLATRGSALYFHIGETEKKCFIEEIPDETMVIGNYRTQLYDKQREEYQPATPGL 62
Query 118 TCSIIFKDPSGRSVYSREVLPADAEGKVTHMTTTTGEY 155
+ KDP + + +R+ +EG+ T + T GE+
Sbjct 63 GMFVEVKDPEDKVILARQY---GSEGRFTFTSHTPGEH 97
> Hs20541596
Length=227
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 41/83 (49%), Gaps = 14/83 (16%)
Query 84 YFYVQESQDKCFIESVPTGVALTASYKNH-----------ENPGVTCSIIFKDPSGRSVY 132
YF++ E++ +CFIE +P + +Y+ PG+ + KDP G+ V
Sbjct 31 YFHIGETEKRCFIEEIPDETMVIGNYRTQMWDKQKEVFLPSTPGLGMHVEVKDPDGKVVL 90
Query 133 SREVLPADAEGKVTHMTTTTGEY 155
SR+ +EG+ T + T G++
Sbjct 91 SRQY---GSEGRFTFTSHTPGDH 110
> CE05883
Length=204
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 33/69 (47%), Gaps = 4/69 (5%)
Query 76 FAHSVSAAYFYVQESQDKCFIESVPTGVALTASYKNHENPGVTCSIIFKDPSGRSVYSRE 135
AHS+ FYV + KC E + V +T Y+ + T SI D G ++Y RE
Sbjct 13 LAHSLR---FYVPPKEKKCLKEEIHKNVVVTGEYEFSQGIQYTGSIHVTDTRGHTLYKRE 69
Query 136 VLPADAEGK 144
AD +GK
Sbjct 70 NF-ADLKGK 77
> SPBC16E9.09c
Length=215
Score = 31.2 bits (69), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 24/46 (52%), Gaps = 5/46 (10%)
Query 82 AAYFYVQESQDKCFIESVPTGVALT-----ASYKNHENPGVTCSII 122
A +FY S+ KCF++ +P G+ +T + N N VT + I
Sbjct 19 AVHFYFDGSKPKCFLKDMPKGMMVTGIVKAEQWNNEANKWVTSNNI 64
> 7292705
Length=210
Score = 31.2 bits (69), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 22/89 (24%), Positives = 36/89 (40%), Gaps = 5/89 (5%)
Query 69 LVLALGYFAHSVSAAYFYVQESQD--KCFIESVPTGVALTASYKNHENPGVTCSIIFKDP 126
+V A+ H +SA F + + CF E + + ++ + + KDP
Sbjct 6 VVFAVALMMHCISAVEFTFDLADNAVDCFYEEIKKNSSAYFEFQVSAGGQLDVDVTLKDP 65
Query 127 SGRSVYSREVLPADAEGKVTHMTTTTGEY 155
G+ +YS E D+ V TTG Y
Sbjct 66 QGKVIYSLEKATFDSHQFVAE---TTGVY 91
> Hs18558120
Length=101
Score = 30.8 bits (68), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 25/51 (49%), Gaps = 0/51 (0%)
Query 81 SAAYFYVQESQDKCFIESVPTGVALTASYKNHENPGVTCSIIFKDPSGRSV 131
+A YF+ E ++KC IE +P+ +T + GV I PS R V
Sbjct 14 TAFYFHAGEREEKCLIEDIPSDTLVTEPWGEGIVVGVQVQIGDCFPSKRLV 64
> CE03255
Length=710
Score = 30.8 bits (68), Expect = 1.0, Method: Composition-based stats.
Identities = 23/105 (21%), Positives = 51/105 (48%), Gaps = 4/105 (3%)
Query 29 RSPVFTFARIVQLLVSFF---LTRAMRTLQRIGGLPPFRAAAALVLALGYFAHSVSAAYF 85
R P F+F ++V+ ++ A ++ + G L R + L++ + + ++ ++F
Sbjct 161 RYPDFSFRQLVETVIQQLEGAFALAFKSSRFPGQLVASRRGSPLLVGIKSNS-TLHTSHF 219
Query 86 YVQESQDKCFIESVPTGVALTASYKNHENPGVTCSIIFKDPSGRS 130
V S+ +CF+ + T + S+ N + SI ++PSG +
Sbjct 220 PVSYSKGRCFMSNNATHLREETSFVETPNNILDLSIAVRNPSGSA 264
> At1g80540
Length=481
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 29/73 (39%), Gaps = 9/73 (12%)
Query 15 WLRRNGHLIATPCRRSPVFTFARIVQLLVSFFLTRAMRTLQRIGGLPPFRAAAALVLALG 74
W N H IA PC +P + R L+ + AM + RIG +A L
Sbjct 87 WNVTNSHYIAPPC--NPSLNYLRTSNNLIRLLICDAMSFIFRIG-------SAMLYTGQN 137
Query 75 YFAHSVSAAYFYV 87
F SV + Y+
Sbjct 138 EFYGSVERTFMYI 150
> 7302459
Length=511
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Query 16 LRRNGHLIATPCRRSPVF---TFARIVQLLVSFFLT 48
+R NG +++ PC RS VF T + + L+ FLT
Sbjct 140 MRHNGEIVSVPCSRSDVFNTKTLTIVEKRLLMKFLT 175
> CE03123
Length=404
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 20/38 (52%), Gaps = 1/38 (2%)
Query 11 DAGSWLRRNGHLIATPCRR-SPVFTFARIVQLLVSFFL 47
DAG W R+N L R PVF A++VQ + F L
Sbjct 85 DAGIWYRKNTTLFFLYHRYIVPVFFIAKVVQFAIPFIL 122
> CE06701
Length=5170
Score = 27.7 bits (60), Expect = 10.0, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 30/64 (46%), Gaps = 2/64 (3%)
Query 77 AHSVSAAYFYVQESQDKCFIESVPTGVALTASYKNHENPGVTCSIIFKDPSGRSVYSREV 136
+H A +V ES +K +V T + T YK H +PG T I+ + P + Y+ V
Sbjct 301 SHDDELAAHFVAESFEKHDEGNVET-IEQTPVYKGHHSPGQTSPIVSEHPHAQE-YAESV 358
Query 137 LPAD 140
D
Sbjct 359 TSHD 362
Lambda K H
0.323 0.136 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2033998736
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40