bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
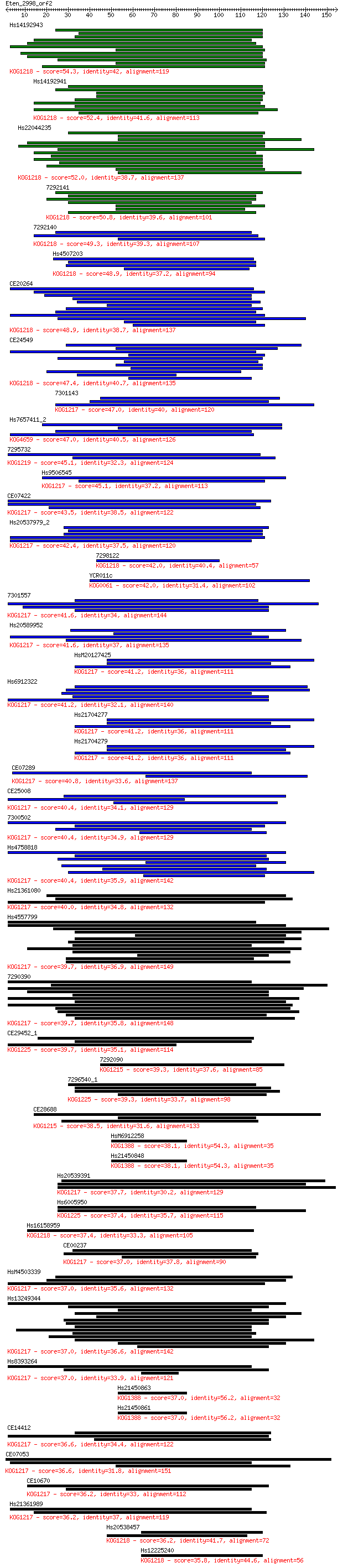
Query= Eten_2998_orf2
Length=154
Score E
Sequences producing significant alignments: (Bits) Value
Hs14192943 54.3 8e-08
Hs14192941 52.4 3e-07
Hs22044235 52.0 4e-07
7292141 50.8 1e-06
7292140 49.3 3e-06
Hs4507203 48.9 4e-06
CE20264 48.9 4e-06
CE24549 47.4 1e-05
7301143 47.0 1e-05
Hs7657411_2 47.0 2e-05
7295732 45.1 6e-05
Hs9506545 45.1 6e-05
CE07422 43.5 2e-04
Hs20537979_2 42.4 3e-04
7298122 42.0 5e-04
YCR011c 42.0 5e-04
7301557 41.6 6e-04
Hs20589952 41.6 6e-04
HsM20127425 41.2 8e-04
Hs6912322 41.2 8e-04
Hs21704277 41.2 8e-04
Hs21704279 41.2 8e-04
CE07289 40.8 0.001
CE25008 40.4 0.001
7300502 40.4 0.001
Hs4758818 40.4 0.001
Hs21361080 40.0 0.002
Hs4557799 39.7 0.002
7290390 39.7 0.002
CE29452_1 39.7 0.002
7292090 39.3 0.003
7296540_1 39.3 0.003
CE28688 38.5 0.005
HsM6912258 38.1 0.007
Hs21450848 38.1 0.007
Hs20539391 37.7 0.009
Hs6005950 37.4 0.012
Hs16158959 37.4 0.012
CE00237 37.0 0.014
HsM4503339 37.0 0.014
Hs13249344 37.0 0.014
Hs8393264 37.0 0.016
Hs21450863 37.0 0.016
Hs21450861 37.0 0.017
CE14412 36.6 0.020
CE07053 36.6 0.020
CE10670 36.2 0.026
Hs21361989 36.2 0.027
Hs20538457 36.2 0.029
Hs12225240 35.8 0.029
> Hs14192943
Length=1140
Score = 54.3 bits (129), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 41/112 (36%), Positives = 50/112 (44%), Gaps = 16/112 (14%)
Query 24 NGVCNSSNTCECTPEFPAANCVDLCSG----------VECSNDGKCDPATGKCTCLAGWG 73
+G C + NTC+C P + NC C G +C N C+P TG C C AG+
Sbjct 115 HGRCIAPNTCQCEPGWGGTNCSSACDGDHWGPHCTSRCQCKNGALCNPITGACHCAAGFR 174
Query 74 GPQCIVPQPCGTEGA------ICSGNATCDQEALSCVCKPGYTGEDCTELSP 119
G +C GT G C ATCD C C PGYTG C +L P
Sbjct 175 GWRCEDRCEQGTYGNDCHQRCQCQNGATCDHVTGECRCPPGYTGAFCEDLCP 226
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 37/91 (40%), Positives = 44/91 (48%), Gaps = 9/91 (9%)
Query 35 CTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSGNA 94
C+P F C +CS C N CD TGKCTC G+ G C P P GT G CS
Sbjct 400 CSPGFYGEACQQICS---CQNGADCDSVTGKCTCAPGFKGIDCSTPCPLGTYGINCSSRC 456
Query 95 TCDQEAL------SCVCKPGYTGEDCTELSP 119
C +A+ SC CK G+ G DC+ P
Sbjct 457 GCKNDAVCSPVDGSCTCKAGWHGVDCSIRCP 487
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 39/103 (37%), Positives = 45/103 (43%), Gaps = 16/103 (15%)
Query 33 CECTPEFPAANCVDLCSGVE----------CSNDGKCDPATGKCTCLAGWGGPQCIVPQP 82
C C P + A C DLC + C N G C TG+C+C +GW G C P P
Sbjct 210 CRCPPGYTGAFCEDLCPPGKHGPQCEQRCPCQNGGVCHHVTGECSCPSGWMGTVCGQPCP 269
Query 83 CGTEGAICS------GNATCDQEALSCVCKPGYTGEDCTELSP 119
G G CS TCD C C PGYTGE C + P
Sbjct 270 EGRFGKNCSQECQCHNGGTCDAATGQCHCSPGYTGERCQDECP 312
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 39/117 (33%), Positives = 51/117 (43%), Gaps = 22/117 (18%)
Query 14 CESDSDCNAPNGVCNSSNTCECTPEFPAANCVDLC----SGVECS------NDGKCDPAT 63
C++ +DC++ G C C P F +C C G+ CS ND C P
Sbjct 415 CQNGADCDSVTG------KCTCAPGFKGIDCSTPCPLGTYGINCSSRCGCKNDAVCSPVD 468
Query 64 GKCTCLAGWGGPQCIVPQPCGTEGAICS------GNATCDQEALSCVCKPGYTGEDC 114
G CTC AGW G C + P GT G C+ C+ +C C PG+ GE C
Sbjct 469 GSCTCKAGWHGVDCSIRCPSGTWGFGCNLTCQCLNGGACNTLDGTCTCAPGWRGEKC 525
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 36/112 (32%), Positives = 47/112 (41%), Gaps = 9/112 (8%)
Query 11 ANCCESDSDCNAPNGVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLA 70
A C D +C G T C F +C +C +C N CD +G+CTC
Sbjct 722 AFCSAYDGECKCTPGWTGLYCTQRCPLGFYGKDCALIC---QCQNGADCDHISGQCTCRT 778
Query 71 GWGGPQCIVPQPCGTEGAICS------GNATCDQEALSCVCKPGYTGEDCTE 116
G+ G C P GT G C N+TCD +C C PG+ G C +
Sbjct 779 GFMGRHCEQKCPSGTYGYGCRQICDCLNNSTCDHITGTCYCSPGWKGARCDQ 830
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 40/133 (30%), Positives = 52/133 (39%), Gaps = 22/133 (16%)
Query 3 GSNCETHEANCCESDSDCNAPNGVCNSSNTCECTPEFPAANCVDLC---------SGV-E 52
G C C S C+ G+C +C P F A C ++C +G+
Sbjct 621 GHRCSQTCPQCVHSSGPCHHITGLC------DCLPGFTGALCNEVCPSGRFGKNCAGICT 674
Query 53 CSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQEAL------SCVCK 106
C+N+G C+P C C GW G C P P G C C A C C
Sbjct 675 CTNNGTCNPIDRSCQCYPGWIGSDCSQPCPPAHWGPNCIHTCNCHNGAFCSAYDGECKCT 734
Query 107 PGYTGEDCTELSP 119
PG+TG CT+ P
Sbjct 735 PGWTGLYCTQRCP 747
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/76 (36%), Positives = 39/76 (51%), Gaps = 7/76 (9%)
Query 52 ECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICS------GNATCDQEALSCVC 105
+C N G CD ATG+C C G+ G +C P GT G +C+ C + +C+C
Sbjct 282 QCHNGGTCDAATGQCHCSPGYTGERCQDECPVGTYGVLCAETCQCVNGGKCYHVSGACLC 341
Query 106 KPGYTGEDC-TELSPE 120
+ G+ GE C L PE
Sbjct 342 EAGFAGERCEARLCPE 357
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 34/118 (28%), Positives = 43/118 (36%), Gaps = 9/118 (7%)
Query 8 THEANCCESDSDCNAPNGVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCT 67
T+ C D C G S + C P NC+ C+ C N C G+C
Sbjct 676 TNNGTCNPIDRSCQCYPGWIGSDCSQPCPPAHWGPNCIHTCN---CHNGAFCSAYDGECK 732
Query 68 CLAGWGGPQCIVPQPCGTEGA------ICSGNATCDQEALSCVCKPGYTGEDCTELSP 119
C GW G C P G G C A CD + C C+ G+ G C + P
Sbjct 733 CTPGWTGLYCTQRCPLGFYGKDCALICQCQNGADCDHISGQCTCRTGFMGRHCEQKCP 790
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 38/115 (33%), Positives = 49/115 (42%), Gaps = 7/115 (6%)
Query 11 ANCCESDSDCNAPNGVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLA 70
A+C D C G ++ C+P F C C S+ G C TG C CL
Sbjct 591 ASCSPDDGICECAPGFRGTTCQRICSPGFYGHRCSQTCPQCVHSS-GPCHHITGLCDCLP 649
Query 71 GWGGPQCIVPQPCGTEGAICSG------NATCDQEALSCVCKPGYTGEDCTELSP 119
G+ G C P G G C+G N TC+ SC C PG+ G DC++ P
Sbjct 650 GFTGALCNEVCPSGRFGKNCAGICTCTNNGTCNPIDRSCQCYPGWIGSDCSQPCP 704
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 33/75 (44%), Gaps = 6/75 (8%)
Query 52 ECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQE------ALSCVC 105
+C N G C+ G CTC GW G +C +P GT G C+ C C C
Sbjct 500 QCLNGGACNTLDGTCTCAPGWRGEKCELPCQDGTYGLNCAERCDCSHADGCHPTTGHCRC 559
Query 106 KPGYTGEDCTELSPE 120
PG++G C + E
Sbjct 560 LPGWSGVHCDSVCAE 574
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 37/130 (28%), Positives = 49/130 (37%), Gaps = 36/130 (27%)
Query 25 GVCNSSN-TCECTP-------EFP------AANCVDLCSGVECSNDGKCDPATGKCTCLA 70
G CN+ + TC C P E P NC + C +CS+ C P TG C CL
Sbjct 505 GACNTLDGTCTCAPGWRGEKCELPCQDGTYGLNCAERC---DCSHADGCHPTTGHCRCLP 561
Query 71 GWGG-------------PQCIVP------QPCGTEGAICSGNATCDQEALSCVCKPGYTG 111
GW G P C +P C + IC +C PG+ G
Sbjct 562 GWSGVHCDSVCAEGRWGPNCSLPCYCKNGASCSPDDGICECAPGFRGTTCQRICSPGFYG 621
Query 112 EDCTELSPEC 121
C++ P+C
Sbjct 622 HRCSQTCPQC 631
Score = 32.0 bits (71), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 35/121 (28%), Positives = 42/121 (34%), Gaps = 18/121 (14%)
Query 18 SDCNAPNG-VCNS-SNTCECTPEFPAANCVDLC----------SGVECSNDGKCDPATGK 65
S C NG +CN + C C F C D C +C N CD TG+
Sbjct 150 SRCQCKNGALCNPITGACHCAAGFRGWRCEDRCEQGTYGNDCHQRCQCQNGATCDHVTGE 209
Query 66 CTCLAGWGGPQCIVPQPCGTEGAIC------SGNATCDQEALSCVCKPGYTGEDCTELSP 119
C C G+ G C P G G C C C C G+ G C + P
Sbjct 210 CRCPPGYTGAFCEDLCPPGKHGPQCEQRCPCQNGGVCHHVTGECSCPSGWMGTVCGQPCP 269
Query 120 E 120
E
Sbjct 270 E 270
> Hs14192941
Length=969
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 38/106 (35%), Positives = 46/106 (43%), Gaps = 16/106 (15%)
Query 30 SNTCECTPEFPAANCVDLCSG----------VECSNDGKCDPATGKCTCLAGWGGPQCIV 79
S CEC P F A C +C+G C+N+G C P G C C GW G C
Sbjct 561 SGICECLPGFSGALCNQVCAGGYFGQDCAQLCSCANNGTCSPIDGSCQCFPGWIGKDCSQ 620
Query 80 PQPCGTEGAI------CSGNATCDQEALSCVCKPGYTGEDCTELSP 119
P G G C A+C E +C C PG+TG CT+ P
Sbjct 621 ACPPGFWGPACFHACSCHNGASCSAEDGACHCTPGWTGLFCTQRCP 666
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 40/112 (35%), Positives = 51/112 (45%), Gaps = 16/112 (14%)
Query 24 NGVCNSSNTCECTPEFPAANCVDLC----------SGVECSNDGKCDPATGKCTCLAGWG 73
+G C S +TC C P + +C C + +C N C+P TG C C AG+
Sbjct 34 HGRCVSPDTCHCEPGWGGPDCSSGCDSDHWGPHCSNRCQCQNGALCNPITGACVCAAGFR 93
Query 74 GPQCIVPQPCGTEGA------ICSGNATCDQEALSCVCKPGYTGEDCTELSP 119
G +C GT G C A+CD A C+C PGYTG C EL P
Sbjct 94 GWRCEELCAPGTHGKGCQLPCQCRHGASCDPRAGECLCAPGYTGVYCEELCP 145
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 34/84 (40%), Positives = 42/84 (50%), Gaps = 9/84 (10%)
Query 43 NCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQEAL- 101
NC +CS C+N G C P G CTC GW G C +P P GT G C+ + TC A
Sbjct 370 NCSSICS---CNNGGTCSPVDGSCTCKEGWQGLDCTLPCPSGTWGLNCNESCTCANGAAC 426
Query 102 -----SCVCKPGYTGEDCTELSPE 120
SC C PG+ G+ C P+
Sbjct 427 SPIDGSCSCTPGWLGDTCELPCPD 450
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 33/83 (39%), Positives = 40/83 (48%), Gaps = 9/83 (10%)
Query 43 NCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICS------GNATC 96
NC + C +CS+ CDP TG C CLAGW G +C P G G CS +C
Sbjct 456 NCSEHC---DCSHADGCDPVTGHCCCLAGWTGIRCDSTCPPGRWGPNCSVSCSCENGGSC 512
Query 97 DQEALSCVCKPGYTGEDCTELSP 119
E SC C PG+ G C + P
Sbjct 513 SPEDGSCECAPGFRGPLCQRICP 535
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 36/103 (34%), Positives = 41/103 (39%), Gaps = 16/103 (15%)
Query 33 CECTPEFPAANCVDLC----SGVEC------SNDGKCDPATGKCTCLAGWGGPQCIVPQP 82
C C P + C +LC G C N G C TG+C C GW G C P P
Sbjct 129 CLCAPGYTGVYCEELCPPGSHGAHCELRCPCQNGGTCHHITGECACPPGWTGAVCAQPCP 188
Query 83 CGTEGAICSGN------ATCDQEALSCVCKPGYTGEDCTELSP 119
GT G CS + CD C C GY G+ C E P
Sbjct 189 PGTFGQNCSQDCPCHHGGQCDHVTGQCHCTAGYMGDRCQEECP 231
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 38/115 (33%), Positives = 51/115 (44%), Gaps = 10/115 (8%)
Query 14 CESDSDCNAPNGVCN---SSNTCECTPEFPAANCVDLCSGV-ECSNDGKCDPATGKCTCL 69
C + + C+A +G C+ CT PAA C V +C N CD +GKCTC
Sbjct 637 CHNGASCSAEDGACHCTPGWTGLFCTQRCPAAFFGKDCGRVCQCQNGASCDHISGKCTCR 696
Query 70 AGWGGPQCIVPQPCGTEGAICS------GNATCDQEALSCVCKPGYTGEDCTELS 118
G+ G C GT G C N+TCD +C C PG+ G C + +
Sbjct 697 TGFTGQHCEQRCAPGTFGYGCQQLCECMNNSTCDHVTGTCYCSPGFKGIRCDQAA 751
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 38/105 (36%), Positives = 46/105 (43%), Gaps = 17/105 (16%)
Query 33 CECTPEFPAANCVDLCS----GVECSND------GKCDPATGKCTCLAGWGGPQCIVPQP 82
C C P + A C C G CS D G+CD TG+C C AG+ G +C P
Sbjct 172 CACPPGWTGAVCAQPCPPGTFGQNCSQDCPCHHGGQCDHVTGQCHCTAGYMGDRCQEECP 231
Query 83 CGTEGAICSGN------ATCDQEALSCVCKPGYTGEDCTE-LSPE 120
G+ G CS C +C C+PGY G C E L PE
Sbjct 232 FGSFGFQCSQRCDCHNGGQCSPTTGACECEPGYKGPRCQERLCPE 276
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 41/138 (29%), Positives = 50/138 (36%), Gaps = 25/138 (18%)
Query 14 CESDSDCNAPNGVCNSSNTCECTPEFPAANCVD-LC----SGVECS--------NDGKCD 60
C DC+ ++ CEC P + C + LC G C+ N C
Sbjct 239 CSQRCDCHNGGQCSPTTGACECEPGYKGPRCQERLCPEGLHGPGCTLPCPCDADNTISCH 298
Query 61 PATGKCTCLAGWGGPQCIVPQPCGTEGA------ICSGNATCDQEALSCVCKPGYTGEDC 114
P TG CTC GW G C P G G C A C C C PG+ GE C
Sbjct 299 PVTGACTCQPGWSGHHCNESCPVGYYGDGCQLPCTCQNGADCHSITGGCTCAPGFMGEVC 358
Query 115 T------ELSPECSGLCT 126
P CS +C+
Sbjct 359 AVSCAAGTYGPNCSSICS 376
Score = 33.5 bits (75), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 37/91 (40%), Gaps = 11/91 (12%)
Query 35 CTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICS--- 91
C P NC CS C N G C P G C C G+ GP C P G G C+
Sbjct 491 CPPGRWGPNCSVSCS---CENGGSCSPEDGSCECAPGFRGPLCQRICPPGFYGHGCAQPC 547
Query 92 -----GNATCDQEALSCVCKPGYTGEDCTEL 117
+ C + C C PG++G C ++
Sbjct 548 PLCVHSSRPCHHISGICECLPGFSGALCNQV 578
> Hs22044235
Length=1229
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 36/108 (33%), Positives = 49/108 (45%), Gaps = 17/108 (15%)
Query 30 SNTCECTPEFPAANCVDLC----------SGVECSNDGKCDPATGKCTCLAGWGGPQCIV 79
+ C C P F + C D+C + C+NDG C PATG C+C GW G C
Sbjct 694 TGACLCLPGFVGSRCQDVCPAGWYGPSCQTRCSCANDGHCHPATGHCSCAPGWTGFSCQR 753
Query 80 PQPCGTEGAICS-------GNATCDQEALSCVCKPGYTGEDCTELSPE 120
G G CS G+ +CD + C+C+ GY G C + P+
Sbjct 754 ACDTGHWGPDCSHPCNCSAGHGSCDAISGLCLCEAGYVGPRCEQQCPQ 801
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 29/73 (39%), Positives = 35/73 (47%), Gaps = 6/73 (8%)
Query 53 CSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSGN------ATCDQEALSCVCK 106
C N G CDP TG+C C AGW G +C P G G C+ A C +C C
Sbjct 900 CLNGGLCDPHTGRCLCPAGWTGDKCQSPCLRGWFGEACAQRCSCPPGAACHHVTGACRCP 959
Query 107 PGYTGEDCTELSP 119
PG+TG C + P
Sbjct 960 PGFTGSGCEQGCP 972
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 33/106 (31%), Positives = 43/106 (40%), Gaps = 21/106 (19%)
Query 53 CSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAI------CSGNATCDQEALSCVCK 106
C N G C + G C+C GW G C + P G GA C N+TC+ +C C
Sbjct 1072 CRNGGLCHASNGSCSCGLGWTGRHCELACPPGRYGAACHLECSCHNNSTCEPATGTCRCG 1131
Query 107 PGYTGEDCT---------------ELSPECSGLCTVGGSVIRDSVS 137
PG+ G+ C + P C+ C GG D VS
Sbjct 1132 PGFYGQACEHRRSGATCNLDCRRGQFGPSCTLHCDCGGGADCDPVS 1177
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 38/116 (32%), Positives = 44/116 (37%), Gaps = 9/116 (7%)
Query 11 ANCCESDSDCNAPNGVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLA 70
A C C P G S C P C LC C N G CD ATG C C
Sbjct 947 AACHHVTGACRCPPGFTGSGCEQGCPPGRYGPGCEQLCG---CLNGGSCDAATGACRCPT 1003
Query 71 GWGGPQCIVPQPCGTEGAICSG------NATCDQEALSCVCKPGYTGEDCTELSPE 120
G+ G C + P G G C+ A CD +C+C PG G C P+
Sbjct 1004 GFLGTDCNLTCPQGRFGPNCTHVCGCGQGAACDPVTGTCLCPPGRAGVRCERGCPQ 1059
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 46/145 (31%), Positives = 54/145 (37%), Gaps = 26/145 (17%)
Query 25 GVCNS-SNTCECTPEFPAANCVDLC----------SGVECSNDGKCDPATGKCTCLAGWG 73
G C+S + C C P NC D C C+N G+C G C C G
Sbjct 469 GTCDSVTGACRCPPGVSGTNCEDGCPKGYYGKHCRKKCNCANRGRCHRLYGACLCDPGLY 528
Query 74 GPQCIVPQPCGTEGAICS--------GNATCDQEALSCVCKPGYTGEDCTE------LSP 119
G C + P G CS +CD+ SC CK G+ GE C P
Sbjct 529 GRFCHLTCPPWAFGPGCSEECQCVQPHTQSCDKRDGSCSCKAGFRGERCQAECELGYFGP 588
Query 120 ECSGLCTVGGSVIRDSVS-ECKVDC 143
C CT V DSVS EC C
Sbjct 589 GCWQACTCPVGVACDSVSGECGKRC 613
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 39/121 (32%), Positives = 48/121 (39%), Gaps = 10/121 (8%)
Query 7 ETHEANCCESDSDCNAPNGVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKC 66
+ H +C + D C+ G EC + C C+ C CD +G+C
Sbjct 553 QPHTQSCDKRDGSCSCKAGFRGERCQAECELGYFGPGCWQACT---CPVGVACDSVSGEC 609
Query 67 --TCLAGWGGPQCIVPQPCGTEGAICS-----GNATCDQEALSCVCKPGYTGEDCTELSP 119
C AG+ G C P GT G CS G A C C C PG TGEDC P
Sbjct 610 GKRCPAGFQGEDCGQECPVGTFGVNCSSSCSCGGAPCHGVTGQCRCPPGRTGEDCEADCP 669
Query 120 E 120
E
Sbjct 670 E 670
Score = 37.4 bits (85), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 36/120 (30%), Positives = 47/120 (39%), Gaps = 17/120 (14%)
Query 14 CESDSDCNAPNGVCNS-SNTCECTPEFPAANCVDLC----------SGVECSNDGKCDPA 62
C +C+A +G C++ S C C + C C +C + CD
Sbjct 764 CSHPCNCSAGHGSCDAISGLCLCEAGYVGPRCEQQCPQGHFGPGCEQRCQCQHGAACDHV 823
Query 63 TGKCTCLAGWGGPQCIVPQPCGTEGAI------CSGNATCDQEALSCVCKPGYTGEDCTE 116
+G CTC AGW G C P G G C+ A CD SC+C G G C E
Sbjct 824 SGACTCPAGWRGTFCEHACPAGFFGLDCRSACNCTAGAACDAVNGSCLCPAGRRGPRCAE 883
Score = 35.8 bits (81), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 35/116 (30%), Positives = 49/116 (42%), Gaps = 18/116 (15%)
Query 22 APNGVCN-SSNTCECTPEFPAANCVDLCS----GVECSN-------DGKCDPATGKCTCL 69
A +G C+ ++ C C P + +C C G +CS+ G CD +G C C
Sbjct 728 ANDGHCHPATGHCSCAPGWTGFSCQRACDTGHWGPDCSHPCNCSAGHGSCDAISGLCLCE 787
Query 70 AGWGGPQCIVPQPCGTEGA------ICSGNATCDQEALSCVCKPGYTGEDCTELSP 119
AG+ GP+C P G G C A CD + +C C G+ G C P
Sbjct 788 AGYVGPRCEQQCPQGHFGPGCEQRCQCQHGAACDHVSGACTCPAGWRGTFCEHACP 843
Score = 35.0 bits (79), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 41/128 (32%), Positives = 50/128 (39%), Gaps = 25/128 (19%)
Query 14 CESDSDCNAPNGVCN-----SSNTCE--CTPEFPAANCVDLCSGVECSNDGKCDPATGKC 66
C + C+A NG C+ + CE C P A C CS C N+ C+PATG C
Sbjct 1072 CRNGGLCHASNGSCSCGLGWTGRHCELACPPGRYGAACHLECS---CHNNSTCEPATGTC 1128
Query 67 TCLAGWGGPQCIVPQ---------------PCGTEGAICSGNATCDQEALSCVCKPGYTG 111
C G+ G C + P T C G A CD + C C GY G
Sbjct 1129 RCGPGFYGQACEHRRSGATCNLDCRRGQFGPSCTLHCDCGGGADCDPVSGQCHCVDGYMG 1188
Query 112 EDCTELSP 119
C E P
Sbjct 1189 PTCREGGP 1196
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 32/100 (32%), Positives = 39/100 (39%), Gaps = 13/100 (13%)
Query 26 VCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGT 85
+CN + C P+ NC CS C N G CD TG C C G G C P G
Sbjct 445 ICNET----CPPDTFGKNCSFSCS---CQNGGTCDSVTGACRCPPGVSGTNCEDGCPKGY 497
Query 86 EGAI------CSGNATCDQEALSCVCKPGYTGEDCTELSP 119
G C+ C + +C+C PG G C P
Sbjct 498 YGKHCRKKCNCANRGRCHRLYGACLCDPGLYGRFCHLTCP 537
Score = 32.3 bits (72), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 37/120 (30%), Positives = 46/120 (38%), Gaps = 20/120 (16%)
Query 20 CNAPNGVCNSSNTCECTPEFPAANCVDLCS--------GVECSND-----GKCDPATGKC 66
C P GV S + EC PA + C GV CS+ C TG+C
Sbjct 594 CTCPVGVACDSVSGECGKRCPAGFQGEDCGQECPVGTFGVNCSSSCSCGGAPCHGVTGQC 653
Query 67 TCLAGWGGPQCIVPQPCGTEGA-------ICSGNATCDQEALSCVCKPGYTGEDCTELSP 119
C G G C P G G C A CD E +C+C PG+ G C ++ P
Sbjct 654 RCPPGRTGEDCEADCPEGRWGLGCQEICPACQHAARCDPETGACLCLPGFVGSRCQDVCP 713
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 31/75 (41%), Gaps = 6/75 (8%)
Query 52 ECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICS------GNATCDQEALSCVC 105
+C N G C C C GW G C P T G CS TCD +C C
Sbjct 421 DCRNGGTCLLGLDGCDCPEGWTGLICNETCPPDTFGKNCSFSCSCQNGGTCDSVTGACRC 480
Query 106 KPGYTGEDCTELSPE 120
PG +G +C + P+
Sbjct 481 PPGVSGTNCEDGCPK 495
Score = 28.1 bits (61), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 34/97 (35%), Gaps = 12/97 (12%)
Query 53 CSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICS------GNATCDQEALSCVCK 106
C C TG C C G+ G C P G G C +CD +C C
Sbjct 943 CPPGAACHHVTGACRCPPGFTGSGCEQGCPPGRYGPGCEQLCGCLNGGSCDAATGACRCP 1002
Query 107 PGYTGEDCT------ELSPECSGLCTVGGSVIRDSVS 137
G+ G DC P C+ +C G D V+
Sbjct 1003 TGFLGTDCNLTCPQGRFGPNCTHVCGCGQGAACDPVT 1039
> 7292141
Length=434
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 38/113 (33%), Positives = 49/113 (43%), Gaps = 17/113 (15%)
Query 24 NGVCN-SSNTCECTPEFPAANCVDLC----------SGVECSNDGKCDPATGKCTCLAGW 72
N VC S CEC + A C D+C C N GKC +G+C C G+
Sbjct 8 NAVCEPFSGDCECAKGYTGARCADICPEGFFGANCSEKCRCENGGKCHHVSGECQCAPGF 67
Query 73 GGPQCIVPQPCGTEGAICS------GNATCDQEALSCVCKPGYTGEDCTELSP 119
GP C + P G GA C + C E +C+C PG+TG+ C P
Sbjct 68 TGPLCDMRCPDGKHGAQCQQDCPCQNDGKCQPETGACMCNPGWTGDVCANKCP 120
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 37/103 (35%), Positives = 41/103 (39%), Gaps = 16/103 (15%)
Query 30 SNTCECTPEFPA----ANCVDLCSGVECS------NDGKCDPATGKCTCLAGWGGPQCIV 79
S C+C P F C D G +C NDGKC P TG C C GW G C
Sbjct 58 SGECQCAPGFTGPLCDMRCPDGKHGAQCQQDCPCQNDGKCQPETGACMCNPGWTGDVCAN 117
Query 80 PQPCGTEGAICS------GNATCDQEALSCVCKPGYTGEDCTE 116
P G+ G C A C C C PGY GE C +
Sbjct 118 KCPVGSYGPGCQESCECYKGAPCHHITGQCECPPGYRGERCFD 160
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 39/115 (33%), Positives = 50/115 (43%), Gaps = 18/115 (15%)
Query 20 CNAPNGV-CNSSN-TCECTPEFPAANCVDLCS----------GVECSNDGKCDPATGKCT 67
CN NG C+ N TC C P + C + C +C N KC+P TG+C
Sbjct 264 CNCKNGAKCSPVNGTCLCAPGWRGPTCEESCEPGTFGQDCALRCDCQNGAKCEPETGQCL 323
Query 68 CLAGWGGPQCIVPQPCGTEGAICS------GNATCDQEALSCVCKPGYTGEDCTE 116
C AGW +C P G C+ NA C+ + SC C G+TGE C
Sbjct 324 CTAGWKNIKCDRPCDLNHFGQDCAKVCDCHNNAACNPQNGSCTCAAGWTGERCER 378
Score = 37.0 bits (84), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 29/101 (28%), Positives = 41/101 (40%), Gaps = 16/101 (15%)
Query 30 SNTCECTPEFPAANCVDLCSGVE----------CSNDGKCDPATGKCTCLAGWGGPQC-- 77
+ C+C+ + +A C C+ + C N KC P G C C GW GP C
Sbjct 233 TGNCQCSIGWSSAQCTRPCTFLRYGPNCELTCNCKNGAKCSPVNGTCLCAPGWRGPTCEE 292
Query 78 -IVPQPCGTEGAI---CSGNATCDQEALSCVCKPGYTGEDC 114
P G + A+ C A C+ E C+C G+ C
Sbjct 293 SCEPGTFGQDCALRCDCQNGAKCEPETGQCLCTAGWKNIKC 333
Score = 35.8 bits (81), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 34/75 (45%), Gaps = 6/75 (8%)
Query 52 ECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICS------GNATCDQEALSCVC 105
+C N+ C+P +G C C G+ G +C P G GA CS C + C C
Sbjct 4 DCLNNAVCEPFSGDCECAKGYTGARCADICPEGFFGANCSEKCRCENGGKCHHVSGECQC 63
Query 106 KPGYTGEDCTELSPE 120
PG+TG C P+
Sbjct 64 APGFTGPLCDMRCPD 78
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 30/68 (44%), Gaps = 8/68 (11%)
Query 52 ECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICS--------GNATCDQEALSC 103
+C N+ C+P G CTC AGW G +C G G C+ + CD C
Sbjct 351 DCHNNAACNPQNGSCTCAAGWTGERCERKCDTGKFGHDCAQKCQCDFNNSLACDATNGRC 410
Query 104 VCKPGYTG 111
VCK + G
Sbjct 411 VCKQDWGG 418
Score = 33.9 bits (76), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 30/106 (28%), Positives = 39/106 (36%), Gaps = 19/106 (17%)
Query 30 SNTCECTPEFPAANCVDLCS----------GVECSNDGKCDPATGKCTCLAGWGGPQCIV 79
+ CEC P + C D C +C+ND CD A G C C GW G +C
Sbjct 144 TGQCECPPGYRGERCFDECQLNTYGFNCSMTCDCANDAMCDRANGTCICNPGWTGAKCAE 203
Query 80 PQ-PCGTEGAICSGNATCDQEAL--------SCVCKPGYTGEDCTE 116
G C+ CD E +C C G++ CT
Sbjct 204 RICEANKYGLDCNRTCECDMEHTDLCHPETGNCQCSIGWSSAQCTR 249
> 7292140
Length=594
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 34/99 (34%), Positives = 43/99 (43%), Gaps = 11/99 (11%)
Query 24 NGVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPC 83
+G C S C+C + C +C C N+ CDP +G C C AGW G C P P
Sbjct 108 HGRCISPEKCKCDHGYGGPACDIIC---RCLNNSSCDPDSGNCICSAGWTGADCAEPCPP 164
Query 84 GTEGA--------ICSGNATCDQEALSCVCKPGYTGEDC 114
G G I GN +CD +C+ GY G C
Sbjct 165 GFYGMECKERCPEILHGNKSCDHITGEILCRTGYIGLTC 203
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 39/121 (32%), Positives = 49/121 (40%), Gaps = 24/121 (19%)
Query 14 CESDSDCNAPNGVCNSSNTCECTPEFPAANCVDLC----------SGVECSNDGKCDPAT 63
CE +CN G C+C P + +NC + C C + C A
Sbjct 222 CEHGGECNHVTG------QCQCLPGWTGSNCNESCPTDTYGQGCAQRCRCVHHKVCRKAD 275
Query 64 GKCTCLAGWGGPQCIVPQPCGTEGAIC-------SGNATCDQEALSCVCKPGYTGEDCTE 116
G C C GW G +C P G G C S N C A CVC+ GYTG++C E
Sbjct 276 GMCICETGWSGTRCDEVCPEGFYGEHCMNTCACPSANFQC-HAAHGCVCRSGYTGDNCDE 334
Query 117 L 117
L
Sbjct 335 L 335
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 36/74 (48%), Gaps = 6/74 (8%)
Query 53 CSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQEAL------SCVCK 106
C + G+C+ TG+C CL GW G C P T G C+ C + C+C+
Sbjct 222 CEHGGECNHVTGQCQCLPGWTGSNCNESCPTDTYGQGCAQRCRCVHHKVCRKADGMCICE 281
Query 107 PGYTGEDCTELSPE 120
G++G C E+ PE
Sbjct 282 TGWSGTRCDEVCPE 295
> Hs4507203
Length=830
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 35/94 (37%), Positives = 46/94 (48%), Gaps = 10/94 (10%)
Query 23 PNGVCN-SSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQ 81
P+G C ++ C+C + A C C+ C G+CDPATG C C GW C P
Sbjct 107 PHGQCEPATGACQCQADRWGARCEFPCA---CGPHGRCDPATGVCHCEPGWWSSTCRRPC 163
Query 82 PCGTEGAICSGNATCDQEALSCVCKPGYTGEDCT 115
C T A C+Q +CVCKPG+ G C+
Sbjct 164 QCNT------AAARCEQATGACVCKPGWWGRRCS 191
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 31/95 (32%), Positives = 44/95 (46%), Gaps = 12/95 (12%)
Query 30 SNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAI 89
S C C P + C C V G+C A+G+CTC G+ G +C +P P G+ G
Sbjct 205 SGRCACRPGWWGPECQQQCECVR----GRCSAASGECTCPPGFRGARCELPCPAGSHGVQ 260
Query 90 CS-------GNATCDQEALSC-VCKPGYTGEDCTE 116
C+ N C + SC C+PG+ G C +
Sbjct 261 CAHSCGRCKHNEPCSPDTGSCESCEPGWNGTQCQQ 295
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 23/61 (37%), Positives = 27/61 (44%), Gaps = 3/61 (4%)
Query 56 DGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQ---EALSCVCKPGYTGE 112
G CD TG C C AG+ GP C P G G CS C + +S C+PG
Sbjct 360 QGSCDTVTGDCVCSAGYWGPSCNASCPAGFHGNNCSVPCECPEGLCHPVSGSCQPGSGSR 419
Query 113 D 113
D
Sbjct 420 D 420
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 34/108 (31%), Positives = 44/108 (40%), Gaps = 20/108 (18%)
Query 29 SSNTCECTPEFPAANCVDLC----SGVECSN-------DGKCDPATGKC-TCLAGWGGPQ 76
+S C C P F A C C GV+C++ + C P TG C +C GW G Q
Sbjct 233 ASGECTCPPGFRGARCELPCPAGSHGVQCAHSCGRCKHNEPCSPDTGSCESCEPGWNGTQ 292
Query 77 CIVPQPCGTEG-------AICSGNATCDQEALSC-VCKPGYTGEDCTE 116
C P GT G C C+ + C C PG+ G C +
Sbjct 293 CQQPCLPGTFGESCEQQCPHCRHGEACEPDTGHCQRCDPGWLGPRCED 340
> CE20264
Length=1111
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 43/125 (34%), Positives = 58/125 (46%), Gaps = 16/125 (12%)
Query 3 GSNCETHEANCCESDSDCNAPNGVCN-----SSNTCE--CTPEFPAANCVDLCSGVECSN 55
G NC H+ C + C+ +G C + + CE C + ANC C +C
Sbjct 467 GPNC-AHQCQCNQRGVGCDGADGKCQCDRGWTGHRCEHHCPADTFGANCEKRC---KCPK 522
Query 56 DGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICS-----GNATCDQEALSCVCKPGYT 110
CDP TG+CTC AG G C + P G+ G C N CD+E C C+PG+
Sbjct 523 GIGCDPITGECTCPAGLQGANCDIGCPEGSYGPGCKLHCKCVNGKCDKETGECTCQPGFF 582
Query 111 GEDCT 115
G DC+
Sbjct 583 GSDCS 587
Score = 41.2 bits (95), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 37/114 (32%), Positives = 45/114 (39%), Gaps = 19/114 (16%)
Query 19 DCNAP--NGVCNSSNTCECTPEFPAANCVDLCS----------GVECSNDGKCDPATGKC 66
DCN P G C CEC P + C CS +C N CDP G C
Sbjct 120 DCNPPCKKGKCIEPGKCECDPGYGGKYCASSCSVGTWGLGCSKSCDCENGANCDPELGTC 179
Query 67 TCLAGWGGPQCIVPQPCGTEGA------ICSGNATCDQEALSCVCKPGYTGEDC 114
C +G+ G +C P P G C C++E CVC G+ GE C
Sbjct 180 ICTSGFQGERCEKPCPDNKWGPNCVKSCPCQNGGKCNKEG-KCVCSDGWGGEFC 232
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 42/123 (34%), Positives = 60/123 (48%), Gaps = 19/123 (15%)
Query 14 CESDSDCNAPNGVC-------NSSNTCECTPEFPAANCVDLCSGVECSNDGK-CDPATGK 65
C S S C+ NG+C + T C+ F C +C C+++ K C+ TG+
Sbjct 735 CASGSTCDHINGLCICPAGLEGALCTRPCSAGFWGNGCRQVC---RCTSEYKQCNAQTGE 791
Query 66 CTCLAGWGGPQCIVPQPCGTEGA------ICSGNAT--CDQEALSCVCKPGYTGEDCTEL 117
C+C AG+ G +C P G G C G AT C++ + +C C PG+TGE C L
Sbjct 792 CSCPAGFQGDRCDKPCEDGYYGPDCIKKCKCQGTATSSCNRVSGACHCHPGFTGEFCHAL 851
Query 118 SPE 120
PE
Sbjct 852 CPE 854
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 36/99 (36%), Positives = 40/99 (40%), Gaps = 17/99 (17%)
Query 32 TCECTPEFPAANCVDLCSGVE----------CSNDGKCDPATGKCTCLAGWGGPQCIVPQ 81
TC CT F C C + C N GKC+ GKC C GWGG C+
Sbjct 178 TCICTSGFQGERCEKPCPDNKWGPNCVKSCPCQNGGKCN-KEGKCVCSDGWGGEFCLNKC 236
Query 82 PCGTEGAICS------GNATCDQEALSCVCKPGYTGEDC 114
G GA C ATCD C+CK GY G C
Sbjct 237 EEGKFGAECKFECNCQNGATCDNTNGKCICKSGYHGALC 275
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 42/97 (43%), Gaps = 16/97 (16%)
Query 34 ECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICS-- 91
EC+ F + C C +C N+ CD ++G+C C+ GW G C + G G C
Sbjct 278 ECSVGFFGSGCTQKC---DCLNNQNCDSSSGECKCI-GWTGKHCDIGCSRGRFGLQCKQN 333
Query 92 ----------GNATCDQEALSCVCKPGYTGEDCTELS 118
NA+CD + C C+ GY G C E
Sbjct 334 CTCPGLEFSDSNASCDAKTGQCQCESGYKGPKCDERK 370
Score = 37.4 bits (85), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 30/78 (38%), Positives = 42/78 (53%), Gaps = 12/78 (15%)
Query 48 CSGVECSN-DGKCDPATGKCTCLAGWGGPQCIVPQPCGTE--GAICSGNATCDQEAL--- 101
C G+E S+ + CD TG+C C +G+ GP+C + C E GA CS TC +E
Sbjct 336 CPGLEFSDSNASCDAKTGQCQCESGYKGPKCD-ERKCDAEQYGADCSKTCTCVRENTLMC 394
Query 102 -----SCVCKPGYTGEDC 114
C CKPG+ G++C
Sbjct 395 APNTGFCRCKPGFYGDNC 412
Score = 36.6 bits (83), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 39/110 (35%), Gaps = 19/110 (17%)
Query 29 SSNTCECTPEFPAANCVDLCS----GVECSNDGKCD--------PATGKCTCLAGWGGPQ 76
++ C C P F NC CS G C CD P TG C C G G
Sbjct 397 NTGFCRCKPGFYGDNCELACSKDSYGPNCEKQAMCDWNHASECNPETGSCVCKPGRTGKN 456
Query 77 CIVPQPCGTEGAICSGNATCDQEAL-------SCVCKPGYTGEDCTELSP 119
C P P G C+ C+Q + C C G+TG C P
Sbjct 457 CSEPCPLDFYGPNCAHQCQCNQRGVGCDGADGKCQCDRGWTGHRCEHHCP 506
Score = 36.2 bits (82), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 36/105 (34%), Positives = 45/105 (42%), Gaps = 12/105 (11%)
Query 24 NGVCNS----SNTCECTPEFPAANCVDLCSGV-ECSNDGKCDPATGKCTCLAGWGGPQCI 78
NGVC S S+ C PA + D C V C++ CDP TG+C C G+ G C
Sbjct 657 NGVCLSCPPGSSGIHCEHNCPAGSYGDGCQQVCSCADGHGCDPTTGECICEPGYHGKTCS 716
Query 79 VPQPCGTEGA-------ICSGNATCDQEALSCVCKPGYTGEDCTE 116
P G G C+ +TCD C+C G G CT
Sbjct 717 EKCPDGKYGYGCALDCPKCASGSTCDHINGLCICPAGLEGALCTR 761
Score = 34.7 bits (78), Expect = 0.067, Method: Compositional matrix adjust.
Identities = 43/132 (32%), Positives = 50/132 (37%), Gaps = 17/132 (12%)
Query 3 GSNCETHEANCCESDSDCNAPNG--VCNSSNTCE-----CTPEFPAANCVDLCSGVECSN 55
G NCE S+CN G VC T + C +F NC C +C+
Sbjct 422 GPNCEKQAMCDWNHASECNPETGSCVCKPGRTGKNCSEPCPLDFYGPNCAHQC---QCNQ 478
Query 56 DG-KCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSGN------ATCDQEALSCVCKPG 108
G CD A GKC C GW G +C P T GA C CD C C G
Sbjct 479 RGVGCDGADGKCQCDRGWTGHRCEHHCPADTFGANCEKRCKCPKGIGCDPITGECTCPAG 538
Query 109 YTGEDCTELSPE 120
G +C PE
Sbjct 539 LQGANCDIGCPE 550
Score = 32.7 bits (73), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 39/137 (28%), Positives = 52/137 (37%), Gaps = 23/137 (16%)
Query 25 GVCNSSNTCECTPEFPAANCVDLCS----GVECS------NDGKCDPATGKCTCLAGWGG 74
G CN C C+ + C++ C G EC N CD GKC C +G+ G
Sbjct 213 GKCNKEGKCVCSDGWGGEFCLNKCEEGKFGAECKFECNCQNGATCDNTNGKCICKSGYHG 272
Query 75 PQCIVPQPCGTEGAICS------GNATCDQEALSCVCKPGYTGEDCT------ELSPECS 122
C G G+ C+ N CD + C C G+TG+ C +C
Sbjct 273 ALCENECSVGFFGSGCTQKCDCLNNQNCDSSSGECKCI-GWTGKHCDIGCSRGRFGLQCK 331
Query 123 GLCTVGGSVIRDSVSEC 139
CT G DS + C
Sbjct 332 QNCTCPGLEFSDSNASC 348
Score = 31.2 bits (69), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 31/66 (46%), Gaps = 5/66 (7%)
Query 56 DGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICS-----GNATCDQEALSCVCKPGYT 110
+GKCD TG+CTC G+ G C G G C +A+C ++ C+C G
Sbjct 565 NGKCDKETGECTCQPGFFGSDCSTTCSKGKYGESCELSCPCSDASCSKQTGKCLCPLGTK 624
Query 111 GEDCTE 116
G C +
Sbjct 625 GVSCDQ 630
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 31/68 (45%), Gaps = 7/68 (10%)
Query 60 DPATGKC-TCLAGWGGPQCIVPQPCGTEGA------ICSGNATCDQEALSCVCKPGYTGE 112
DP G C +C G G C P G+ G C+ CD C+C+PGY G+
Sbjct 654 DPKNGVCLSCPPGSSGIHCEHNCPAGSYGDGCQQVCSCADGHGCDPTTGECICEPGYHGK 713
Query 113 DCTELSPE 120
C+E P+
Sbjct 714 TCSEKCPD 721
> CE24549
Length=1664
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 31/87 (35%), Positives = 42/87 (48%), Gaps = 12/87 (13%)
Query 52 ECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSG------NATCDQEALSCVC 105
C+N CD +G+C C G+ G C P GT G CS N+ CD + C C
Sbjct 1258 RCANSKSCDHISGRCQCPKGYAGHSCTELCPDGTFGESCSQKCDCGENSMCDAISGKCFC 1317
Query 106 KPGYTGEDCT------ELSPECSGLCT 126
KPG++G DC P+C+ LC+
Sbjct 1318 KPGHSGSDCKSGCVQGRFGPDCNQLCS 1344
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 41/131 (31%), Positives = 52/131 (39%), Gaps = 22/131 (16%)
Query 29 SSNTCECTPEFPAANCVDLC----SGVEC------SNDGKCDPATGKCTCLAGWGGPQCI 78
S +C CTP F A C ++C G++C N CD + G C C GW G +C
Sbjct 871 SDGSCHCTPGFYGATCSEVCPTGRFGIDCMQLCKCQNGAICDTSNGSCECAPGWSGKKCD 930
Query 79 VPQPCGTEGAICSGN------ATCDQEALSCVCKPGYTGEDCTE------LSPECSGLCT 126
GT G CS CD C+C PG G C E C G+C+
Sbjct 931 KACAPGTFGKDCSKKCDCADGMHCDPSDGECICPPGKKGHKCDETCDSGLFGAGCKGICS 990
Query 127 VGGSVIRDSVS 137
DSV+
Sbjct 991 CQNGATCDSVT 1001
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 43/127 (33%), Positives = 52/127 (40%), Gaps = 18/127 (14%)
Query 3 GSNCETHEANCCESDSDCNAPNGVCN-----SSNTCECTPEF--PAANCVDLCSGVECSN 55
GSNC H C +CN NG C + +CE F NC C+ C N
Sbjct 1121 GSNCMKHCL--CMHGGECNKENGDCECIDGWTGPSCEFLCPFGQFGRNCAQRCN---CKN 1175
Query 56 DGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQEAL------SCVCKPGY 109
CD TG+C CL GW G C G GA C C+ AL C C+PG+
Sbjct 1176 GASCDRKTGRCECLPGWSGEHCEKSCVSGHYGAKCEETCECENGALCDPISGHCSCQPGW 1235
Query 110 TGEDCTE 116
G+ C
Sbjct 1236 RGKKCNR 1242
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 27/69 (39%), Positives = 34/69 (49%), Gaps = 6/69 (8%)
Query 58 KCDPATGKCTCLAGWGGPQCIVPQPCGTEGA------ICSGNATCDQEALSCVCKPGYTG 111
+CD TG+C C AGW GP C P G G CS A+CD+ C C G+ G
Sbjct 1049 RCDHVTGECRCPAGWTGPDCQTSCPLGRHGEGCRHSCQCSNGASCDRVTGFCDCPSGFMG 1108
Query 112 EDCTELSPE 120
++C PE
Sbjct 1109 KNCESECPE 1117
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 37/111 (33%), Positives = 46/111 (41%), Gaps = 16/111 (14%)
Query 25 GVCNSSN-TCECTPEFPAANCVDLCSGVE----------CSNDGKCDPATGKCTCLAGWG 73
GVC+SS +C C P + C C C N G CD TG+C CL G+
Sbjct 1349 GVCDSSTGSCVCPPGYIGTKCEIACQSDRFGPTCEKICNCENGGTCDRLTGQCRCLPGFT 1408
Query 74 GPQCIVPQPCGTEGAICS-----GNATCDQEALSCVCKPGYTGEDCTELSP 119
G C P G GA C N C+ + C C G+TG C + P
Sbjct 1409 GMTCNQVCPEGRFGAGCKEKCRCANGHCNASSGECKCNLGFTGPSCEQSCP 1459
Score = 41.2 bits (95), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 31/70 (44%), Gaps = 8/70 (11%)
Query 56 DGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICS--------GNATCDQEALSCVCKP 107
+G+CDP G CTC G G C P P T G C + CD+ C CKP
Sbjct 683 NGRCDPVFGYCTCPDGLYGQSCEKPCPHFTFGKNCRFPCKCARENSEGCDEITGKCRCKP 742
Query 108 GYTGEDCTEL 117
GY G C +
Sbjct 743 GYYGHHCKRM 752
Score = 36.2 bits (82), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 34/74 (45%), Gaps = 6/74 (8%)
Query 52 ECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGA------ICSGNATCDQEALSCVC 105
+CSN CD TG C C +G+ G C P G G+ +C C++E C C
Sbjct 1086 QCSNGASCDRVTGFCDCPSGFMGKNCESECPEGLWGSNCMKHCLCMHGGECNKENGDCEC 1145
Query 106 KPGYTGEDCTELSP 119
G+TG C L P
Sbjct 1146 IDGWTGPSCEFLCP 1159
Score = 36.2 bits (82), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 26/67 (38%), Positives = 31/67 (46%), Gaps = 6/67 (8%)
Query 59 CDPATGKCTCLAGWGGPQCIVPQPCGTEGAICS------GNATCDQEALSCVCKPGYTGE 112
C TG CTCL G GP C T G C+ A CD+ SC C PG+ G
Sbjct 825 CHHVTGTCTCLPGKTGPLCDQSCAPNTYGPNCAHTCSCVNGAKCDESDGSCHCTPGFYGA 884
Query 113 DCTELSP 119
C+E+ P
Sbjct 885 TCSEVCP 891
Score = 34.3 bits (77), Expect = 0.089, Method: Compositional matrix adjust.
Identities = 36/107 (33%), Positives = 42/107 (39%), Gaps = 17/107 (15%)
Query 20 CNAPNGVCN-SSNTCECTPEFPAANCVDLC-SGV---------ECSNDGKCDPATGKCTC 68
C NG CN SS C+C F +C C SG EC +CDP G C C
Sbjct 1429 CRCANGHCNASSGECKCNLGFTGPSCEQSCPSGKYGLNCTLDCECYGQARCDPVQGCCDC 1488
Query 69 LAGWGGPQCIVPQPCGTEGAICS------GNATCDQEALSCVCKPGY 109
G G +C P G G CS A CD C+C G+
Sbjct 1489 PPGRYGSRCQFSCPNGFYGWYCSQSCSCQNGAHCDGADGRCLCPAGF 1535
Score = 33.9 bits (76), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 24/46 (52%), Gaps = 3/46 (6%)
Query 34 ECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIV 79
EC P +C + C +C N CD TG+C+C GW GP C +
Sbjct 1598 ECRPGRYGQSCQNKC---QCFNGATCDARTGQCSCSPGWLGPTCQI 1640
Score = 33.1 bits (74), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 24/63 (38%), Positives = 25/63 (39%), Gaps = 6/63 (9%)
Query 58 KCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSGN------ATCDQEALSCVCKPGYTG 111
KCD GKC C G GP C G G C ATCD C C PG+ G
Sbjct 1576 KCDATDGKCICPVGRHGPLCEEECRPGRYGQSCQNKCQCFNGATCDARTGQCSCSPGWLG 1635
Query 112 EDC 114
C
Sbjct 1636 PTC 1638
> 7301143
Length=2146
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 40/85 (47%), Gaps = 3/85 (3%)
Query 45 VDLCSGVECSNDGKCDPATG--KCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQEALS 102
VD C C N G+C G C CL GWGG C P C T+ + G
Sbjct 426 VDECDKNPCLNGGRCFDTYGWYTCQCLDGWGGEICDRPMTCQTQQCLNGGTCLDKPIGFQ 485
Query 103 CVCKPGYTGEDCTELSPECSGLCTV 127
C+C P YTGE C +++P C+ C +
Sbjct 486 CLCPPEYTGELC-QIAPSCAQQCPI 509
Score = 33.1 bits (74), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 30/89 (33%), Positives = 43/89 (48%), Gaps = 10/89 (11%)
Query 40 PAANCVDLCSGVECSNDGKCDPATGK--CTCLAGWGGPQC--IVPQPCGTEGAICSGNAT 95
P C+ LC +C NDG C + + CTC G+ G C + + TE C N T
Sbjct 1753 PEEGCI-LCFQSDCKNDGFCQSPSDEYACTCQPGFEGDDCGTDIDECLNTE---CLNNGT 1808
Query 96 CDQE--ALSCVCKPGYTGEDCTELSPECS 122
C + A C C+PG+ G+ C + EC+
Sbjct 1809 CINQVAAFFCQCQPGFEGQHCEQNIDECA 1837
Score = 32.7 bits (73), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 44/132 (33%), Positives = 55/132 (41%), Gaps = 15/132 (11%)
Query 24 NGVCNS---SNTCECTPEFPAANC---VDLCSGVECSNDGKCDPATG--KCTCLAGWGGP 75
NG+C + S C CTP F +C VD C C N C +C C G+ G
Sbjct 659 NGICKNEKGSYKCYCTPGFTGVHCDSDVDECLSFPCLNGATCHNKINAYECVCQPGYEGE 718
Query 76 QCIVP-QPCGTEGAICSGNATCDQEA--LSCVCKPGYTGEDCTELSPECSG-LCTVGGSV 131
C V CG+ CS +TC +C C PG TG C +C G C GG
Sbjct 719 NCEVDIDECGSNP--CSNGSTCIDRINNFTCNCIPGMTGRICDIDIDDCVGDPCLNGGQC 776
Query 132 IRDSVSECKVDC 143
I D + + DC
Sbjct 777 I-DQLGGFRCDC 787
> Hs7657411_2
Length=2326
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 44/116 (37%), Positives = 57/116 (49%), Gaps = 12/116 (10%)
Query 18 SDCNAPNGVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQC 77
++CN NG C S + C C P F +C V C +G+ + G C C GW GP+C
Sbjct 135 TNCNG-NGECISGH-CHCFPGFLGPDCARDSCPVLCGGNGEYE--KGHCVCRHGWKGPEC 190
Query 78 IVPQPCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTE---LSPECS--GLCTVG 128
VP+ + C G+ TC C+C PGY GE C E L P CS G+C G
Sbjct 191 DVPEEQCID-PTCFGHGTCIMGV--CICVPGYKGEICEEEDCLDPMCSNHGICVKG 243
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 36/80 (45%), Positives = 43/80 (53%), Gaps = 8/80 (10%)
Query 53 CSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQEALSCVCKPGYTGE 112
CSN G C G+C C GWGG C P P E CSG+ T +A C C P +TG
Sbjct 234 CSNHGIC--VKGECHCSTGWGGVNCETPLPVCQEQ--CSGHGTFLLDAGVCSCDPKWTGS 289
Query 113 DC-TEL-SPEC--SGLCTVG 128
DC TEL + EC G+C+ G
Sbjct 290 DCSTELCTMECGSHGVCSRG 309
Score = 34.7 bits (78), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 41/97 (42%), Gaps = 9/97 (9%)
Query 24 NGVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPC 83
+GVC S C+C + C + C+ G+C GKC C GW G C +
Sbjct 303 HGVC-SRGICQCEEGWVGPTCEERSCHSHCTEHGQC--KDGKCECSPGWEGDHCTIAHYL 359
Query 84 GTEG----AICSGNA--TCDQEALSCVCKPGYTGEDC 114
+C GN T DQ CVC+ G++G C
Sbjct 360 DAVRDGCPGLCFGNGRCTLDQNGWHCVCQVGWSGTGC 396
Score = 32.7 bits (73), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 36/114 (31%), Positives = 53/114 (46%), Gaps = 13/114 (11%)
Query 3 GSNCETHEANCCESDSDCNAPNGVCNSSNTCECTPEFPAANC-VDLCSGVECSNDGKCDP 61
G NCET C E C+ + C C P++ ++C +LC+ +EC + G C
Sbjct 253 GVNCETPLPVCQEQ---CSGHGTFLLDAGVCSCDPKWTGSDCSTELCT-MECGSHGVC-- 306
Query 62 ATGKCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQEALSCVCKPGYTGEDCT 115
+ G C C GW GP C + C + C+ + C C C PG+ G+ CT
Sbjct 307 SRGICQCEEGWVGPTC-EERSCHSH---CTEHGQCKDG--KCECSPGWEGDHCT 354
> 7295732
Length=5147
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 36/135 (26%), Positives = 44/135 (32%), Gaps = 27/135 (20%)
Query 2 SGSNCETHEANC----CESDSDCNAPNGVCNSSNTCECTPEFPAANC----VDLCSGVEC 53
SG C + C C S C S C C +C D+C C
Sbjct 4006 SGEQCSRRQDPCLPNPCHSQVQCRR----LGSDFQCMCPANRDGKHCEKERSDVCYSKPC 4061
Query 54 SNDGKC----DPATGKCTCLAGWGGPQC------IVPQPCGTEGAICSGNATCDQEALSC 103
N G C D ++ C C G+ G QC P PC G +C + C
Sbjct 4062 RNGGSCQRSPDGSSYFCLCRPGFRGNQCESVSDSCRPNPC-LHGGLC----VSLKPGYKC 4116
Query 104 VCKPGYTGEDCTELS 118
C PG G C S
Sbjct 4117 NCTPGRYGRHCERFS 4131
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 30/113 (26%), Positives = 43/113 (38%), Gaps = 22/113 (19%)
Query 32 TCECTPEFPAANCV---DLCSGVECSNDGKCDP--ATGKCTCLAGWGGPQC-------IV 79
+C+CT F C D C C + +C + +C C A G C
Sbjct 3998 SCQCTSGFSGEQCSRRQDPCLPNPCHSQVQCRRLGSDFQCMCPANRDGKHCEKERSDVCY 4057
Query 80 PQPCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTELSPEC-------SGLC 125
+PC G S + D + C+C+PG+ G C +S C GLC
Sbjct 4058 SKPCRNGG---SCQRSPDGSSYFCLCRPGFRGNQCESVSDSCRPNPCLHGGLC 4107
> Hs9506545
Length=685
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 42/117 (35%), Positives = 52/117 (44%), Gaps = 6/117 (5%)
Query 18 SDCNAPNGVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQC 77
S C+ NG C+ C C P + C + C + G C +CTC GWGG C
Sbjct 224 SGCHEQNGYCSKPAECLCRPGWQGRLCNECIPHNGCRH-GTCS-TPWQCTCDEGWGGLFC 281
Query 78 IVPQPCGTEGAICSGNATCD---QEALSCVCKPGYTGEDCTELSPEC-SGLCTVGGS 130
T + C ATC Q + +C C+PGYTG DC EC S C GGS
Sbjct 282 DQDLNYCTHHSPCKNGATCSNSGQRSYTCTCRPGYTGVDCELELSECDSNPCRNGGS 338
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 27/90 (30%), Positives = 35/90 (38%), Gaps = 12/90 (13%)
Query 35 CTPEFPAANCVDLCSGV-ECSNDGKCDPATGKCTCLAGWGGPQCIVP---QPCGTEGAIC 90
C+ + NC LC + C P G +CL GW G C P C + C
Sbjct 175 CSDNYYGDNCSRLCKKRNDHFGHYVCQP-DGNLSCLPGWTGEYCQQPICLSGCHEQNGYC 233
Query 91 SGNATCDQEALSCVCKPGYTGEDCTELSPE 120
S A C+C+PG+ G C E P
Sbjct 234 SKPA-------ECLCRPGWQGRLCNECIPH 256
> CE07422
Length=372
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 47/143 (32%), Positives = 66/143 (46%), Gaps = 27/143 (18%)
Query 2 SGSNCETHEANCCESDSDCNAPNGVC---NSSNTCECTPEFPAANCVD----LCSGVECS 54
+GS CE C+S DC++ NG+C S TC C + C +C +C+
Sbjct 199 TGSRCEKTPVALCDS-RDCSS-NGLCIGTKDSMTCACYLGYSGDKCEKITGTMCEASDCN 256
Query 55 NDGKC--DPATGKCTCLAGWGGPQC-----IVPQPCGTEGAIC-----SGNATCDQEAL- 101
++G C C C G+ G +C ++P GTEG C +GN C L
Sbjct 257 SNGICIGTKNLKSCICAPGYYGSRCESRFALLP---GTEGMFCEAKDCNGNGICIGNKLL 313
Query 102 -SCVCKPGYTGEDCTELSPECSG 123
C+C PG+TG C EL P C+G
Sbjct 314 PMCICAPGFTGLRC-ELEPLCTG 335
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 40/133 (30%), Positives = 59/133 (44%), Gaps = 21/133 (15%)
Query 2 SGSNCETHEANCCESDSDCNAPNGVC---NSSNTCECTPEFPAANCVD-----------L 47
SG CE CE+ SDCN+ NG+C + +C C P + + C
Sbjct 238 SGDKCEKITGTMCEA-SDCNS-NGICIGTKNLKSCICAPGYYGSRCESRFALLPGTEGMF 295
Query 48 CSGVECSNDGKC--DPATGKCTCLAGWGGPQCIVPQPCGTEGAICSGNATC--DQEALSC 103
C +C+ +G C + C C G+ G +C + +P T CSGN C ++ SC
Sbjct 296 CEAKDCNGNGICIGNKLLPMCICAPGFTGLRCEL-EPLCTGALQCSGNGLCIGSLKSYSC 354
Query 104 VCKPGYTGEDCTE 116
C G+TG C +
Sbjct 355 TCNLGWTGPTCAQ 367
Score = 31.6 bits (70), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 27/103 (26%), Positives = 47/103 (45%), Gaps = 5/103 (4%)
Query 21 NAPNGVCNSSNT-CECTPEFPAANCVDLCSGVECSNDGKC--DPATGKCTCLAGWGGPQC 77
AP +CN T +C + + CS +C+N G C + C C G+ G +C
Sbjct 144 KAPLCLCNLGKTGFKCEMNMDPSAPITFCSPSDCNNKGLCLGTKNSFSCACQIGYTGSRC 203
Query 78 IVPQPCGTEGAICSGNATC--DQEALSCVCKPGYTGEDCTELS 118
+ CS N C +++++C C GY+G+ C +++
Sbjct 204 EKTPVALCDSRDCSSNGLCIGTKDSMTCACYLGYSGDKCEKIT 246
> Hs20537979_2
Length=1158
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 36/101 (35%), Positives = 45/101 (44%), Gaps = 6/101 (5%)
Query 28 NSSNTCECTPEFPAANC---VDLCSGVECSNDGKCDPATG--KCTCLAGWGGPQCIVPQ- 81
N S TC C F C V+ C+ C N G C +C C AG+GGP C Q
Sbjct 37 NPSYTCSCLSGFTGRRCHLDVNECASQPCQNGGTCTHGINSFRCQCPAGFGGPTCETAQS 96
Query 82 PCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTELSPECS 122
PC T+ G + + CVC+ GYTG C +CS
Sbjct 97 PCDTKECQHGGQCQVENGSAVCVCQAGYTGAACEMDVDDCS 137
Score = 36.2 bits (82), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 35/99 (35%), Positives = 44/99 (44%), Gaps = 11/99 (11%)
Query 30 SNTCECTPEFPAANC---VDLCSGVECSNDGKCDPATGK--CTCLAGWGGPQC-IVPQPC 83
+ C C F +C VD C C + G+C+ G C C + G C V PC
Sbjct 557 AYVCRCPAGFVGVHCETEVDACDSSPCQHGGRCESGGGAYLCVCPESFFGYHCETVSDPC 616
Query 84 GTEGAICSGNATC--DQEALSCVCKPGYTGEDCT-ELSP 119
+ + C G C + SC CK GYTGEDC EL P
Sbjct 617 FS--SPCGGRGYCLASNGSHSCTCKVGYTGEDCAKELFP 653
Score = 35.8 bits (81), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 32/100 (32%), Positives = 43/100 (43%), Gaps = 9/100 (9%)
Query 28 NSSNTCECTPEFPAANC----VDLCSGVECSNDGKCDPATGK--CTCLAGWGGPQCIVPQ 81
+ S TCEC F +C LCS C N G C A G+ C+C + G C + +
Sbjct 305 DDSYTCECPRGFHGKHCEKARPHLCSSGPCRNGGTCKEAGGEYHCSCPYRFTGRHCEIGK 364
Query 82 PCGTEGAICSGNATCDQE--ALSCVCKPGYTGEDCTELSP 119
P C TC C C PG++G C E++P
Sbjct 365 PDSCASGPCHNGGTCFHYIGKYKCDCPPGFSGRHC-EIAP 403
Score = 32.7 bits (73), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 42/132 (31%), Positives = 51/132 (38%), Gaps = 18/132 (13%)
Query 3 GSNCETHEANC----CESDSDCNAPNGVCNSSNTCECTPEFPAANC---VDLCSGVECSN 55
G CET ++ C C+ C NG S C C + A C VD CS C N
Sbjct 88 GPTCETAQSPCDTKECQHGGQCQVENG----SAVCVCQAGYTGAACEMDVDDCSPDPCLN 143
Query 56 DGKCDPATGKCTCL--AGWGGPQCIV---PQPCGTEGAICSGNATC--DQEALSCVCKPG 108
G C G TCL + G +C P P A C TC + C C G
Sbjct 144 GGSCVDLVGNYTCLCAEPFKGLRCETGDHPVPDACLSAPCHNGGTCVDADQGYVCECPEG 203
Query 109 YTGEDCTELSPE 120
+ G DC E P+
Sbjct 204 FMGLDCRERVPD 215
Score = 32.0 bits (71), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 36/122 (29%), Positives = 44/122 (36%), Gaps = 12/122 (9%)
Query 3 GSNCETHEANCCESDSDCNAPNGVCNSSN---TCECTPEFPAANC----VDLCSGVECSN 55
G +CE + C S N G C + C C F +C D C+ C N
Sbjct 318 GKHCEKARPHLCSSGPCRNG--GTCKEAGGEYHCSCPYRFTGRHCEIGKPDSCASGPCHN 375
Query 56 DGKCDPATGK--CTCLAGWGGPQC-IVPQPCGTEGAICSGNATCDQEALSCVCKPGYTGE 112
G C GK C C G+ G C I P PC + G C C+ GY G
Sbjct 376 GGTCFHYIGKYKCDCPPGFSGRHCEIAPSPCFRSPCVNGGTCEDRDTDFFCHCQAGYMGR 435
Query 113 DC 114
C
Sbjct 436 RC 437
> 7298122
Length=832
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 23/57 (40%), Positives = 32/57 (56%), Gaps = 3/57 (5%)
Query 43 NCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQE 99
+C +LC +C N CD +G C C+AGW G C +P P GT G +C CD++
Sbjct 28 DCKNLC---QCQNGAACDNKSGLCHCIAGWTGQFCELPCPQGTYGIMCRKACDCDEK 81
> YCR011c
Length=1049
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 32/115 (27%), Positives = 44/115 (38%), Gaps = 16/115 (13%)
Query 40 PAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQE 99
P NC + EC +C+ TG+C C+ G+ G C +P CG SGN
Sbjct 55 PCFNC--MLPIFECKQFSECNSYTGRCECIEGFAGDDCSLPL-CGGLSPDESGNKDRPIR 111
Query 100 AL--SCVCKPGYTGEDCTELS-----------PECSGLCTVGGSVIRDSVSECKV 141
A +C C G+ G +C P G C G ++ S C V
Sbjct 112 AQNDTCHCDNGWGGINCDVCQEDFVCDAFMPDPSIKGTCYKNGMIVDKVFSGCNV 166
> 7301557
Length=1404
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 34/98 (34%), Positives = 41/98 (41%), Gaps = 20/98 (20%)
Query 33 CECTPEFPAANC---VDLCSGVECSNDGKCDPATG--KCTCLAGWGGPQCIV------PQ 81
C C P + C +D C G C N G C+ G +C C G+ GP C + PQ
Sbjct 861 CRCAPGWTGLFCAEAIDQCRGQPCHNGGTCESGAGWFRCVCAQGFSGPDCRINVNECSPQ 920
Query 82 PCGTEGAICSGNATCDQE--ALSCVCKPGYTGEDCTEL 117
PC G ATC SC+C PG G C L
Sbjct 921 PC-------QGGATCIDGIGGYSCICPPGRHGLRCEIL 951
Score = 32.7 bits (73), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 43/149 (28%), Positives = 59/149 (39%), Gaps = 12/149 (8%)
Query 2 SGSNCETHEANCCESDSDCNAPNGVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDP 61
G NCE EA C + C+ +G C+ CEC P + C + C G C+
Sbjct 275 QGVNCE--EAIC---KAGCDPVHGKCDRPGECECRPGWRGPLCNE-CMVYPGCKHGSCNG 328
Query 62 ATGKCTCLAGWGGPQCIVP-QPCGTEGAICSGNATCDQEA---LSCVCKPGYTGEDCTEL 117
+ KC C WGG C CGT C TC+ A C C G +GE C +
Sbjct 329 SAWKCVCDTNWGGILCDQDLNFCGTHEP-CKHGGTCENTAPDKYRCTCAEGLSGEQCEIV 387
Query 118 SPECSGL-CTVGGSVIRDSVSECKVDCYS 145
C+ C GG+ + + + Y
Sbjct 388 EHPCATRPCRNGGTCTLKTSNRTQAQVYR 416
Score = 32.3 bits (72), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 31/101 (30%), Positives = 39/101 (38%), Gaps = 17/101 (16%)
Query 33 CECTPEFPAANC---VDLCSGVECSNDGKCDPATGK--CTCLAGWGGPQCI------VPQ 81
C C F +C +D C+ C N G+C GK C C G+ G C P
Sbjct 630 CACASGFKGRDCETDIDECATSPCRNGGECVDMVGKFNCICPLGYSGSLCEEAKENCTPS 689
Query 82 PCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTELSPECS 122
PC G+ E C C P G+ C +L P CS
Sbjct 690 PC------LEGHCLNTPEGYYCHCPPDRAGKHCEQLRPLCS 724
Score = 32.3 bits (72), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 40/134 (29%), Positives = 52/134 (38%), Gaps = 22/134 (16%)
Query 9 HEANCCESDSDCNAPN-----GVC---NSSNTCECTPEFPAANCVDLCSG---VECSNDG 57
H CE + + +PN G+C + TCEC + C + +G +C N G
Sbjct 787 HTGTFCEHNLNECSPNPCRNGGICLDGDGDFTCECMSGWTGKRCSERATGCYAGQCQNGG 846
Query 58 KCDPATG------KCTCLAGWGGPQCI-VPQPCGTEGAICSGNATCDQEA--LSCVCKPG 108
C P C C GW G C C G C TC+ A CVC G
Sbjct 847 TCMPGAPDKALQPHCRCAPGWTGLFCAEAIDQC--RGQPCHNGGTCESGAGWFRCVCAQG 904
Query 109 YTGEDCTELSPECS 122
++G DC ECS
Sbjct 905 FSGPDCRINVNECS 918
> Hs20589952
Length=737
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 39/104 (37%), Positives = 47/104 (45%), Gaps = 19/104 (18%)
Query 31 NTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQC-IVPQPCGTEGAI 89
N C P AA C DL +G EC CLA + G C + PC
Sbjct 507 NECLSAPCLNAATCRDLVNGYEC-------------VCLAEYKGTHCELYKDPCAN--VS 551
Query 90 CSGNATCDQEAL--SCVCKPGYTGEDCTELSPEC-SGLCTVGGS 130
C ATCD + L +C+C PG+TGE+C EC S C GGS
Sbjct 552 CLNGATCDSDGLNGTCICAPGFTGEECDIDINECDSNPCHHGGS 595
Score = 34.3 bits (77), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 32/73 (43%), Gaps = 9/73 (12%)
Query 51 VECSNDGKC----DPATGKCTCLAGWGGPQCI-----VPQPCGTEGAICSGNATCDQEAL 101
+ECS GKC AT CTC + G C +PC + N D
Sbjct 317 LECSGKGKCTTKPSEATFSCTCEEQYVGTFCEEYDACQRKPCQNNASCIDANEKQDGSNF 376
Query 102 SCVCKPGYTGEDC 114
+CVC PGYTGE C
Sbjct 377 TCVCLPGYTGELC 389
Score = 32.3 bits (72), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 40/132 (30%), Positives = 58/132 (43%), Gaps = 14/132 (10%)
Query 3 GSNCETHEA---NCCESDSDCNAPNGVCNSSN-TCECTPEFPAANC---VDLCSGVECSN 55
G+ CE ++A C++++ C N + SN TC C P + C +D C C N
Sbjct 344 GTFCEEYDACQRKPCQNNASCIDANEKQDGSNFTCVCLPGYTGELCQSKIDYCILDPCRN 403
Query 56 DGKCDPATG--KCTCLAGWGGPQCIVP-QPCGTEGAICSGNATC--DQEALSCVCKPGYT 110
C + C C G+ G C PC + + C N TC D +C C PG+T
Sbjct 404 GATCISSLSGFTCQCPEGYFGSACEEKVDPCAS--SPCQNNGTCYVDGVHFTCNCSPGFT 461
Query 111 GEDCTELSPECS 122
G C +L C+
Sbjct 462 GPTCAQLIDFCA 473
Score = 31.2 bits (69), Expect = 0.92, Method: Compositional matrix adjust.
Identities = 33/114 (28%), Positives = 44/114 (38%), Gaps = 5/114 (4%)
Query 29 SSNTCECTPEFPAANC---VDLCSGVECSNDGKC--DPATGKCTCLAGWGGPQCIVPQPC 83
S TC+C + + C VD C+ C N+G C D C C G+ GP C
Sbjct 412 SGFTCQCPEGYFGSACEEKVDPCASSPCQNNGTCYVDGVHFTCNCSPGFTGPTCAQLIDF 471
Query 84 GTEGAICSGNATCDQEALSCVCKPGYTGEDCTELSPECSGLCTVGGSVIRDSVS 137
G + C+C PGY G C E EC + + RD V+
Sbjct 472 CALSPCAHGTCRSVGTSYKCLCDPGYHGLYCEEEYNECLSAPCLNAATCRDLVN 525
> HsM20127425
Length=1238
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 33/105 (31%), Positives = 47/105 (44%), Gaps = 14/105 (13%)
Query 48 CSGVECSNDGKCDPA--TGKCTCLAGWGGPQCIV-------PQPCGTEGAICSGNATCDQ 98
C CSN G C + T +C C GW G C V P PC G C G+
Sbjct 716 CDAYTCSNGGTCYDSGDTFRCACPPGWKGSTCAVAKNSSCLPNPC-VNGGTCVGSGA--- 771
Query 99 EALSCVCKPGYTGEDCTELSPECSGLCTVGGSVIRDSVSECKVDC 143
+ SC+C+ G+ G CT + +C+ L G + D V+ + +C
Sbjct 772 -SFSCICRDGWEGRTCTHNTNDCNPLPCYNGGICVDGVNWFRCEC 815
Score = 28.9 bits (63), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 33/79 (41%), Gaps = 3/79 (3%)
Query 48 CSGVECSNDGKCD--PATGKCTCLAGWGGPQCIVP-QPCGTEGAICSGNATCDQEALSCV 104
C+ C+N G C P+ +C C +GW GP C + C + G + C+
Sbjct 351 CTSNPCANGGSCHEVPSGFECHCPSGWSGPTCALDIDECASNPCAAGGTCVDQVDGFECI 410
Query 105 CKPGYTGEDCTELSPECSG 123
C + G C + EC G
Sbjct 411 CPEQWVGATCQLDANECEG 429
Score = 28.1 bits (61), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 40/131 (30%), Positives = 49/131 (37%), Gaps = 31/131 (23%)
Query 33 CECTPEFPAANC---VDLCSGVECSNDGKCDPATG--KCTCLAGWGGPQCIVPQ-PC--- 83
C C F C VDLC C N +C G C C +GG C VP+ PC
Sbjct 522 CHCPQGFSGPLCEVDVDLCEPSPCRNGARCYNLEGDYYCACPDDFGGKNCSVPREPCPGG 581
Query 84 --------------GTEGAICSG----NATCDQEA---LSCVCKPGYTGEDCTELSPECS 122
G G SG + C + SC+C G+TG C E +C
Sbjct 582 ACRVIDGCGSDAGPGMPGTAASGVCGPHGRCVSQPGGNFSCICDSGFTGTYCHENIDDCL 641
Query 123 GL-CTVGGSVI 132
G C GG+ I
Sbjct 642 GQPCRNGGTCI 652
> Hs6912322
Length=1376
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 36/121 (29%), Positives = 54/121 (44%), Gaps = 13/121 (10%)
Query 33 CECTPEFPAANC---VDLCSGVECSNDGKCDPATG--KCTCLAGWGGPQC----IVPQPC 83
C C P + NC +D C +C+N C T C C + G C + C
Sbjct 1200 CTCEPGYTGVNCEVDIDNCQSHQCANGATCISHTNGYSCLCFGNFTGKFCRQSRLPSTVC 1259
Query 84 GTEGAICS----GNATCDQEALSCVCKPGYTGEDCTELSPECSGLCTVGGSVIRDSVSEC 139
G E + GN T Q L C+C+PG+TGE C + EC+ V G + +D +++
Sbjct 1260 GNEKTNLTCYNGGNCTEFQTELKCMCRPGFTGEWCEKDIDECASDPCVNGGLCQDLLNKF 1319
Query 140 K 140
+
Sbjct 1320 Q 1320
Score = 34.3 bits (77), Expect = 0.086, Method: Compositional matrix adjust.
Identities = 41/133 (30%), Positives = 48/133 (36%), Gaps = 21/133 (15%)
Query 29 SSNTCECTPEFPAANC---VDLCSGVECSNDGKCDPATGKCTC--------LAGWGGPQC 77
S C C P F +C V+ CS C N G C+ G TC +GG C
Sbjct 379 SGYVCICQPGFTGIHCEEDVNECSSNPCQNGGTCENLPGNYTCHCPFDNLSRTFYGGRDC 438
Query 78 IVPQPCGTEGAICSGNATC------DQEALSCVCKPGYTGEDC---TELSPECSGLCTVG 128
G C N TC Q SC+C GYTG C T LS E G V
Sbjct 439 -SDILLGCTHQQCLNNGTCIPHFQDGQHGFSCLCPSGYTGSLCEIATTLSFEGDGFLWVK 497
Query 129 GSVIRDSVSECKV 141
+ S C +
Sbjct 498 SGSVTTKGSVCNI 510
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 34/89 (38%), Gaps = 15/89 (16%)
Query 27 CNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQC-IVPQPCGT 85
C N C P NC D+ S CS C GW G C + C +
Sbjct 1137 CLQLNVCNSNPCLHGGNCEDIYSSYHCS-------------CPLGWSGKHCELNIDECFS 1183
Query 86 EGAICSGNATCDQEALSCVCKPGYTGEDC 114
I GN + A C C+PGYTG +C
Sbjct 1184 NPCI-HGNCSDRVAAYHCTCEPGYTGVNC 1211
Score = 30.4 bits (67), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 32/105 (30%), Positives = 46/105 (43%), Gaps = 22/105 (20%)
Query 32 TCECTPEFPAANC---VDLCSGVECSNDGKCDPATGKCTCL--AGWGGPQCIV------P 80
+C C P + +C VD C+ C N+ C G+ TC+ + G C +
Sbjct 171 SCFCVPGYQGRHCDLEVDECASDPCKNEATCLNEIGRYTCICPHNYSGVNCELEIDECWS 230
Query 81 QPCGTEGAICSGNATCDQEAL---SCVCKPGYTGEDCTELSPECS 122
QPC GA C Q+AL C C PG+ G+ C + EC+
Sbjct 231 QPC-LNGATC-------QDALGAYFCDCAPGFLGDHCELNTDECA 267
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 32/124 (25%), Positives = 51/124 (41%), Gaps = 18/124 (14%)
Query 2 SGSNCETHEANCCESDSDCNAPNGVCNSSNTCECTPEFPAANC-VDL--CSGVECSNDGK 58
+G++CET C NA + TC C P + A C +DL C+ C ++G+
Sbjct 294 TGTHCETLMPLCWSKPCHNNATCEDSVDNYTCHCWPGYTGAQCEIDLNECNSNPCQSNGE 353
Query 59 CDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTELS 118
C + + + G + S + + C+C+PG+TG C E
Sbjct 354 CVELSSE---------------KQYGRITGLPSSFSYHEASGYVCICQPGFTGIHCEEDV 398
Query 119 PECS 122
ECS
Sbjct 399 NECS 402
> Hs21704277
Length=1238
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 33/105 (31%), Positives = 47/105 (44%), Gaps = 14/105 (13%)
Query 48 CSGVECSNDGKCDPA--TGKCTCLAGWGGPQCIV-------PQPCGTEGAICSGNATCDQ 98
C CSN G C + T +C C GW G C V P PC G C G+
Sbjct 716 CDAYTCSNGGTCYDSGDTFRCACPPGWKGSTCAVAKNSSCLPNPC-VNGGTCVGSGA--- 771
Query 99 EALSCVCKPGYTGEDCTELSPECSGLCTVGGSVIRDSVSECKVDC 143
+ SC+C+ G+ G CT + +C+ L G + D V+ + +C
Sbjct 772 -SFSCICRDGWEGRTCTHNTNDCNPLPCYNGGICVDGVNWFRCEC 815
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 33/79 (41%), Gaps = 3/79 (3%)
Query 48 CSGVECSNDGKCD--PATGKCTCLAGWGGPQCIVP-QPCGTEGAICSGNATCDQEALSCV 104
C+ C+N G C P+ +C C +GW GP C + C + G + C+
Sbjct 351 CTSNPCANGGSCHEVPSGFECHCPSGWSGPTCALDIDECASNPCAAGGTCVDQVDGFECI 410
Query 105 CKPGYTGEDCTELSPECSG 123
C + G C + EC G
Sbjct 411 CPEQWVGATCQLDANECEG 429
Score = 28.1 bits (61), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 40/131 (30%), Positives = 49/131 (37%), Gaps = 31/131 (23%)
Query 33 CECTPEFPAANC---VDLCSGVECSNDGKCDPATG--KCTCLAGWGGPQCIVPQ-PC--- 83
C C F C VDLC C N +C G C C +GG C VP+ PC
Sbjct 522 CHCPQGFSGPLCEVDVDLCEPSPCRNGARCYNLEGDYYCACPDDFGGKNCSVPREPCPGG 581
Query 84 --------------GTEGAICSG----NATCDQEA---LSCVCKPGYTGEDCTELSPECS 122
G G SG + C + SC+C G+TG C E +C
Sbjct 582 ACRVIDGCGSDAGPGMPGTAASGVCGPHGRCVSQPGGNFSCICDSGFTGTYCHENIDDCL 641
Query 123 GL-CTVGGSVI 132
G C GG+ I
Sbjct 642 GQPCRNGGTCI 652
> Hs21704279
Length=1200
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 33/105 (31%), Positives = 47/105 (44%), Gaps = 14/105 (13%)
Query 48 CSGVECSNDGKCDPA--TGKCTCLAGWGGPQCIV-------PQPCGTEGAICSGNATCDQ 98
C CSN G C + T +C C GW G C V P PC G C G+
Sbjct 678 CDAYTCSNGGTCYDSGDTFRCACPPGWKGSTCAVAKNSSCLPNPC-VNGGTCVGSGA--- 733
Query 99 EALSCVCKPGYTGEDCTELSPECSGLCTVGGSVIRDSVSECKVDC 143
+ SC+C+ G+ G CT + +C+ L G + D V+ + +C
Sbjct 734 -SFSCICRDGWEGRTCTHNTNDCNPLPCYNGGICVDGVNWFRCEC 777
Score = 32.0 bits (71), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 24/86 (27%), Positives = 36/86 (41%), Gaps = 3/86 (3%)
Query 48 CSGVECSNDGKCD--PATGKCTCLAGWGGPQCIVP-QPCGTEGAICSGNATCDQEALSCV 104
C+ C+N G C P+ +C C +GW GP C + C + G + C+
Sbjct 351 CTSNPCANGGSCHEVPSGFECHCPSGWSGPTCALDIDECASNPCAAGGTCVDQVDGFECI 410
Query 105 CKPGYTGEDCTELSPECSGLCTVGGS 130
C + G C +C G C GG+
Sbjct 411 CPEQWVGATCQLDVNDCRGQCQHGGT 436
Score = 28.1 bits (61), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 40/131 (30%), Positives = 49/131 (37%), Gaps = 31/131 (23%)
Query 33 CECTPEFPAANC---VDLCSGVECSNDGKCDPATG--KCTCLAGWGGPQCIVPQ-PC--- 83
C C F C VDLC C N +C G C C +GG C VP+ PC
Sbjct 484 CHCPQGFSGPLCEVDVDLCEPSPCRNGARCYNLEGDYYCACPDDFGGKNCSVPREPCPGG 543
Query 84 --------------GTEGAICSG----NATCDQEA---LSCVCKPGYTGEDCTELSPECS 122
G G SG + C + SC+C G+TG C E +C
Sbjct 544 ACRVIDGCGSDAGPGMPGTAASGVCGPHGRCVSQPGGNFSCICDSGFTGTYCHENIDDCL 603
Query 123 GL-CTVGGSVI 132
G C GG+ I
Sbjct 604 GQPCRNGGTCI 614
> CE07289
Length=616
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 38/128 (29%), Positives = 58/128 (45%), Gaps = 17/128 (13%)
Query 4 SNCETHEA---NCCESDSDCNAPNGVCNSSNTCECTPEFPAANCVD---LCSGVECSNDG 57
+NCET ++ N CE+ C + +S+ TC+CTP++ C + C C N G
Sbjct 397 TNCETFDSCYINKCENGGTCVPTYNLLDSTFTCQCTPDWKGTYCEEERYYCDETPCENGG 456
Query 58 KCD-----PATGKCTCLAGWGGPQCIVP-QPCGTEGAICSG---NATCDQE--ALSCVCK 106
+C+ P + CTC W G C + C + +C +ATC + CVC
Sbjct 457 ECEDIIGPPNSYNCTCTPQWTGFNCTIDVDECVEDTTLCKTKDPDATCVNTNGSYYCVCS 516
Query 107 PGYTGEDC 114
P G+ C
Sbjct 517 PNMFGKSC 524
Score = 37.0 bits (84), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 36/85 (42%), Gaps = 15/85 (17%)
Query 66 CTCLAGWGGPQCIV-------PQPCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTELS 118
CTC G+ G C + PQPC G S T SC C G+ GE C
Sbjct 84 CTCAVGYTGTDCTMLTSDPCSPQPCLQNGVCSSSGGT-----YSCACATGFFGEQCQYSG 138
Query 119 PECSGLCTVGGS---VIRDSVSECK 140
CSG C+ GG+ + D+ C+
Sbjct 139 DPCSGYCSNGGTCEMLFSDTTPYCQ 163
> CE25008
Length=2972
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 34/110 (30%), Positives = 50/110 (45%), Gaps = 7/110 (6%)
Query 28 NSSNTCECTPEFPAANCVDLC-----SGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQ- 81
N + CEC F + C ++C + + C N G C+ T C C+ + G C +
Sbjct 1866 NGTVFCECYDGFTSTRCDEVCDVCKANILLCQNSGTCNSTTQSCDCIDYFSGTYCERNEN 1925
Query 82 PCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTELSPECSGL-CTVGGS 130
C T +C TC+ + CVC P YTG+ C CS + C GG+
Sbjct 1926 LCETNLVVCKNGGTCNPKTGLCVCLPDYTGDYCDNQIHSCSDINCFNGGT 1975
Score = 34.7 bits (78), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 41/87 (47%), Gaps = 6/87 (6%)
Query 2 SGSNCETHEANCCESDSDCNAPNGVCN-SSNTCECTPEFPAANC---VDLCSGVECSNDG 57
SG+ CE +E N CE++ G CN + C C P++ C + CS + C N G
Sbjct 1916 SGTYCERNE-NLCETNLVVCKNGGTCNPKTGLCVCLPDYTGDYCDNQIHSCSDINCFNGG 1974
Query 58 KCDPATGKCTCLAGWGGPQC-IVPQPC 83
C C CL G G +C + QPC
Sbjct 1975 TCIDYNATCACLPGTTGDRCQYLGQPC 2001
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 28/90 (31%), Positives = 41/90 (45%), Gaps = 14/90 (15%)
Query 51 VECSNDGKC-DPATG--KCTCLAGWGGPQCI------VPQPCGTEG--AICSGNAT--CD 97
+ C N KC D T +C C G+ G C +PC G +I + N T
Sbjct 1730 LYCQNSVKCFDTFTNYSRCQCAPGFSGDYCSDLIDDCFFEPCFNGGTCSIFNYNTTTKVS 1789
Query 98 QEALSCVCKPGYTGEDC-TELSPECSGLCT 126
E+ +C C+ GY G +C + + P C+ L T
Sbjct 1790 IESYNCTCQTGYFGTNCESRIHPSCTDLIT 1819
> 7300502
Length=833
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 45/146 (30%), Positives = 60/146 (41%), Gaps = 30/146 (20%)
Query 2 SGSNCETHEANCCESDSDCNAPN-----GVCNSSNTCECTPEFPAANC---VDLCSGVEC 53
SG NC+ NC +PN G C S C C F C +D C G +C
Sbjct 411 SGPNCDLQLDNC--------SPNPCINGGSCQPSGKCICPAGFSGTRCETNIDDCLGHQC 462
Query 54 SNDGKCDPATG--KCTCLAGWGGPQCI------VPQPCGTEGAICSGNATCDQEALSCVC 105
N G C +C C+ G+ G C + +PC G + N C C
Sbjct 463 ENGGTCIDMVNQYRCQCVPGFHGTHCSSKVDLCLIRPCANGGTCLNLN-----NDYQCTC 517
Query 106 KPGYTGEDCTELSPEC-SGLCTVGGS 130
+ G+TG+DC+ EC SG C GG+
Sbjct 518 RAGFTGKDCSVDIDECSSGPCHNGGT 543
Score = 37.0 bits (84), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 33/95 (34%), Positives = 43/95 (45%), Gaps = 8/95 (8%)
Query 33 CECTPEFPAANC---VDLCSGVECSNDGKCDPATG--KCTCLAGWGGPQCIVPQPCGTEG 87
C+C P F +C VDLC C+N G C +CTC AG+ G C V + G
Sbjct 477 CQCVPGFHGTHCSSKVDLCLIRPCANGGTCLNLNNDYQCTCRAGFTGKDCSVDIDECSSG 536
Query 88 AICSGNATCDQE--ALSCVCKPGYTGEDCTELSPE 120
C TC + CVC G+ G+ C E S +
Sbjct 537 P-CHNGGTCMNRVNSFECVCANGFRGKQCDEESYD 570
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 38/94 (40%), Gaps = 5/94 (5%)
Query 24 NGVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPC 83
+G C+ N C C + A C + C + P T C C GWGG C
Sbjct 237 HGHCDKPNQCVCQLGWKGALCNECVLEPNCIHGTCNKPWT--CICNEGWGGLYCNQDLNY 294
Query 84 GTEGAICSGNATC---DQEALSCVCKPGYTGEDC 114
T C TC + +C C PGY+G+DC
Sbjct 295 CTNHRPCKNGGTCFNTGEGLYTCKCAPGYSGDDC 328
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 30/62 (48%), Gaps = 10/62 (16%)
Query 63 TGKCTCLAGWGGPQCIVPQ-PCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTE--LSP 119
TG+ CL GW G C +P+ G E + CD+ CVC+ G+ G C E L P
Sbjct 212 TGEIICLTGWQGDYCHIPKCAKGCE------HGHCDKPN-QCVCQLGWKGALCNECVLEP 264
Query 120 EC 121
C
Sbjct 265 NC 266
> Hs4758818
Length=2003
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 49/147 (33%), Positives = 65/147 (44%), Gaps = 27/147 (18%)
Query 2 SGSNCETH----EANCCESDSDCNA-PNGVCNSSNTCECTPEFPAANC---VDLCSGVEC 53
GS C+ H E+ C++ + C A P+G C+C P + NC +D C C
Sbjct 923 QGSLCQDHVNPCESRPCQNGATCMAQPSGY-----LCQCAPGYDGQNCSKELDACQSQPC 977
Query 54 SNDGKCDPATG--KCTCLAGWGGPQCI------VPQPCGTEG-AICSGNATCDQEALSCV 104
N G C P G C C G+ G +C + QPC G A C A A C
Sbjct 978 HNHGTCTPKPGGFHCACPPGFVGLRCEGDVDECLDQPCHPTGTAACHSLAN----AFYCQ 1033
Query 105 CKPGYTGEDC-TELSPECSGLCTVGGS 130
C PG+TG+ C E+ P S C GG+
Sbjct 1034 CLPGHTGQWCEVEIDPCHSQPCFHGGT 1060
Score = 36.2 bits (82), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 39/96 (40%), Gaps = 7/96 (7%)
Query 33 CECTPEFPAANCV----DLCSGVECSNDGKCD-PATGK--CTCLAGWGGPQCIVPQPCGT 85
C C P F C D C CS G+C A+G+ C+C+ GW G QC + C
Sbjct 103 CTCLPGFTGERCQAKLEDPCPPSFCSKRGRCHIQASGRPQCSCMPGWTGEQCQLRDFCSA 162
Query 86 EGAICSGNATCDQEALSCVCKPGYTGEDCTELSPEC 121
+ G + C C PG+ G C EC
Sbjct 163 NPCVNGGVCLATYPQIQCHCPPGFEGHACERDVNEC 198
Score = 35.4 bits (80), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 35/108 (32%), Positives = 41/108 (37%), Gaps = 10/108 (9%)
Query 25 GVCNSS---NTCECTPEFPAANC---VDLCSGVECSNDGKC--DPATGKCTCLAGWGGPQ 76
G CN S C C P C D C C N G C P T C C G+ GP+
Sbjct 742 GSCNPSPGGYYCTCPPSHTGPQCQTSTDYCVSAPCFNGGTCVNRPGTFSCLCAMGFQGPR 801
Query 77 CIVPQPCGTEGAICSGNATCDQ--EALSCVCKPGYTGEDCTELSPECS 122
C + C ATC + C+C GYTG C L C+
Sbjct 802 CEGKLRPSCADSPCRNRATCQDSPQGPRCLCPTGYTGGSCQTLMDLCA 849
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 27/68 (39%), Positives = 33/68 (48%), Gaps = 4/68 (5%)
Query 66 CTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQE--ALSCVCKPGYTGEDCTELSPEC-S 122
C C GW GP+C + G A C+ TC + +C C GYTG C+E C S
Sbjct 677 CVCDVGWTGPECEA-ELGGCISAPCAHGGTCYPQPSGYNCTCPTGYTGPTCSEEMTACHS 735
Query 123 GLCTVGGS 130
G C GGS
Sbjct 736 GPCLNGGS 743
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 30/115 (26%), Positives = 45/115 (39%), Gaps = 25/115 (21%)
Query 27 CNSSNTCECTPEFPAANC------------VDLCSGVECSNDGKC--DPATGKCTCLAGW 72
C + TC+ +P+ P C +DLC+ C + C + C CL GW
Sbjct 815 CRNRATCQDSPQGPRCLCPTGYTGGSCQTLMDLCAQKPCPRNSHCLQTGPSFHCLCLQGW 874
Query 73 GGPQCIVPQPCGTEGAICSG---NATCDQEAL--------SCVCKPGYTGEDCTE 116
GP C +P + A+ G ++ C L C C PG+ G C +
Sbjct 875 TGPLCNLPLSSCQKAALSQGIDVSSLCHNGGLCVDSGPSYFCHCPPGFQGSLCQD 929
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 28/91 (30%), Positives = 37/91 (40%), Gaps = 20/91 (21%)
Query 46 DLCSGVECSNDGKC--DPATGK--CTCLAGWGGPQCIV-----------PQPCGTEGAIC 90
D+C C D +C +P TG C C G+ GP C P PC G+
Sbjct 394 DMCLSQPCHGDAQCSTNPLTGSTLCLCQPGYSGPTCHQDLDECLMAQQGPSPCEHGGSCL 453
Query 91 SGNATCDQEALSCVCKPGYTGEDCTELSPEC 121
+ + +C+C PGYTG C EC
Sbjct 454 NTPGS-----FNCLCPPGYTGSRCEADHNEC 479
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 42/138 (30%), Positives = 55/138 (39%), Gaps = 27/138 (19%)
Query 30 SNTCECTPEFPAANCV--DLCSGVE-CSNDGKCD---PA-------------TGKCTCLA 70
TC+C P F C D C + C N G C PA + CTCL
Sbjct 48 QGTCQCAPGFLGETCQFPDPCQNAQLCQNGGSCQALLPAPLGLPSSPSPLTPSFLCTCLP 107
Query 71 GWGGPQC--IVPQPCGTEGAICSGNATCDQEA---LSCVCKPGYTGEDCTELSPECSGLC 125
G+ G +C + PC + CS C +A C C PG+TGE C +L CS
Sbjct 108 GFTGERCQAKLEDPCPP--SFCSKRGRCHIQASGRPQCSCMPGWTGEQC-QLRDFCSANP 164
Query 126 TVGGSVIRDSVSECKVDC 143
V G V + + + C
Sbjct 165 CVNGGVCLATYPQIQCHC 182
Score = 27.7 bits (60), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 31/63 (49%), Gaps = 10/63 (15%)
Query 65 KCTCLAGWGGPQC-IVPQPCGTEGAICSGNATC------DQEALSCVCKPGYTGEDCTEL 117
+C C G GP+C + PC G CS TC D C+C PG+ G DC E+
Sbjct 219 QCLCPVGQEGPRCELRAGPCPPRG--CSNGGTCQLMPEKDSTFHLCLCPPGFIGPDC-EV 275
Query 118 SPE 120
+P+
Sbjct 276 NPD 278
> Hs21361080
Length=383
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 40/115 (34%), Positives = 49/115 (42%), Gaps = 12/115 (10%)
Query 20 CNAPNGVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIV 79
CN NG C N C C P + C D C G C G+C C GW G C
Sbjct 30 CNPQNGFCEDDNVCRCQPGWQGPLC-DQCVTSPGCLHGLCG-EPGQCICTDGWDGELCDR 87
Query 80 P-QPCGTEGAICSGNATC---DQEALSCVCKPGYTGEDCTELSPECSGLCTVGGS 130
+ C + A C+ N TC D C C PGY+G+DC + G C + GS
Sbjct 88 DVRACSS--APCANNGTCVSLDDGLYECSCAPGYSGKDCQKK----DGPCVINGS 136
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 40/129 (31%), Positives = 56/129 (43%), Gaps = 19/129 (14%)
Query 24 NGVCNSSNTCECTPEFPAANC---VDLCSGVECSNDGKC---DPATGKCTCLAGWGGPQC 77
+G+C C CT + C V CS C+N+G C D +C+C G+ G C
Sbjct 65 HGLCGEPGQCICTDGWDGELCDRDVRACSSAPCANNGTCVSLDDGLYECSCAPGYSGKDC 124
Query 78 IVPQ-PCGTEGAICSGNATC-DQEAL----SCVCKPGYTGEDCTELSPECS-------GL 124
PC G+ C TC D E SC+C PG++G C ++ C+ G+
Sbjct 125 QKKDGPCVINGSPCQHGGTCVDDEGRASHASCLCPPGFSGNFCEIVANSCTPNPCENDGV 184
Query 125 CTVGGSVIR 133
CT G R
Sbjct 185 CTDIGGDFR 193
Score = 31.2 bits (69), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 41/136 (30%), Positives = 56/136 (41%), Gaps = 19/136 (13%)
Query 2 SGSNCETHEANCCESDSDCNAPNGVC------NSSNTCECTPEFPAANC---VDLCSGVE 52
SG +C+ + C + S C G C S +C C P F C + C+
Sbjct 120 SGKDCQKKDGPCVINGSPCQH-GGTCVDDEGRASHASCLCPPGFSGNFCEIVANSCTPNP 178
Query 53 CSNDGKCDPATG--KCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQE---ALSCVCKP 107
C NDG C G +C C AG+ C P + C TC Q + C+CKP
Sbjct 179 CENDGVCTDIGGDFRCRCPAGFIDKTCSRPV-TNCASSPCQNGGTCLQHTQVSYECLCKP 237
Query 108 GYTGEDCTE---LSPE 120
+TG C + LSP+
Sbjct 238 EFTGLTCVKKRALSPQ 253
> Hs4557799
Length=2321
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 45/131 (34%), Positives = 59/131 (45%), Gaps = 23/131 (17%)
Query 2 SGSNC-ETHEANCCESDSDCNAPNGVCNSSNT---CECTPEFPAANCVDL--CSGVECSN 55
SG C ++ + CES C A G C+S C C P C L C+ C +
Sbjct 725 SGPRCSQSLARDACESQP-CRA-GGTCSSDGMGFHCTCPPGVQGRQCELLSPCTPNPCEH 782
Query 56 DGKCDPATGK---CTCLAGWGGPQCIV-------PQPCGTEGAICSGNATCDQEALSCVC 105
G+C+ A G+ C+C GW GP+C P PCG G IC+ A + SC C
Sbjct 783 GGRCESAPGQLPVCSCPQGWQGPRCQQDVDECAGPAPCGPHG-ICTNLAG----SFSCTC 837
Query 106 KPGYTGEDCTE 116
GYTG C +
Sbjct 838 HGGYTGPSCDQ 848
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 48/147 (32%), Positives = 63/147 (42%), Gaps = 21/147 (14%)
Query 2 SGSNCETHEANCCESDSDCNAPNGVCNSSN---TCECTPEFPAANC---VDLCSGVECSN 55
+G++C+ HEA+ C S + GVC++++ C C F C VD CS C N
Sbjct 955 TGAHCQ-HEADPCLSRPCLHG--GVCSAAHPGFRCTCLESFTGPQCQTLVDWCSRQPCQN 1011
Query 56 DGKCDPATGKCTCLAGWGGPQC-IVPQPCGTEGA--------ICSGNATCDQEALS--CV 104
G+C C C GW G C I PC A +C C E S CV
Sbjct 1012 GGRCVQTGAYCLCPPGWSGRLCDIRSLPCREAAAQIGVRLEQLCQAGGQCVDEDSSHYCV 1071
Query 105 CKPGYTGEDC-TELSPECSGLCTVGGS 130
C G TG C E+ P + C GG+
Sbjct 1072 CPEGRTGSHCEQEVDPCLAQPCQHGGT 1098
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 42/146 (28%), Positives = 61/146 (41%), Gaps = 24/146 (16%)
Query 23 PNGVCNS---SNTCECTPEFPAANC---VDLCSGVECSNDGKCDPATGK--CTCLAGWGG 74
P+G+C + S +C C + +C ++ C C N G C G C+CL G+ G
Sbjct 822 PHGICTNLAGSFSCTCHGGYTGPSCDQDINDCDPNPCLNGGSCQDGVGSFSCSCLPGFAG 881
Query 75 PQCI------VPQPCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTELSPECSGLCTVG 128
P+C + PCG G T + +C C PGY G C + P+CS
Sbjct 882 PRCARDVDECLSNPCG------PGTCTDHVASFTCTCPPGYGGFHCEQDLPDCSPSSCFN 935
Query 129 GSVIRDSVSE----CKVDCYSACCKH 150
G D V+ C+ A C+H
Sbjct 936 GGTCVDGVNSFSCLCRPGYTGAHCQH 961
Score = 36.2 bits (82), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 37/115 (32%), Positives = 50/115 (43%), Gaps = 12/115 (10%)
Query 33 CECTPEFPAANC---VDLCS-GVECSNDGKC--DPATGKCTCLAGWGGPQCIVPQ-PCGT 85
C C P + +C VD C G C + G C P + +C C AG+ GP C P PC
Sbjct 144 CSCPPGYQGRSCRSDVDECRVGEPCRHGGTCLNTPGSFRCQCPAGYTGPLCENPAVPCAP 203
Query 86 EGAICSGNATCDQEA---LSCVCKPGYTGEDCTELSPECSGLCTVGGSVIRDSVS 137
+ C TC Q C C PG+ G++C +C G + G D V+
Sbjct 204 --SPCRNGGTCRQSGDLTYDCACLPGFEGQNCEVNVDDCPGHRCLNGGTCVDGVN 256
Score = 35.4 bits (80), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 29/73 (39%), Gaps = 3/73 (4%)
Query 61 PATGKCTCLAGWGGPQC---IVPQPCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTEL 117
P +C C GW GP+C + C ++ G + D C C PG G C L
Sbjct 713 PGGFRCVCEPGWSGPRCSQSLARDACESQPCRAGGTCSSDGMGFHCTCPPGVQGRQCELL 772
Query 118 SPECSGLCTVGGS 130
SP C GG
Sbjct 773 SPCTPNPCEHGGR 785
Score = 34.3 bits (77), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 37/113 (32%), Positives = 45/113 (39%), Gaps = 8/113 (7%)
Query 33 CECTPEFPAANC---VDLCSGVECSNDGKCDPA--TGKCTCLAGWGGPQCIVP-QPCGTE 86
C C P F NC VD C G C N G C T C C W G C C +
Sbjct 222 CACLPGFEGQNCEVNVDDCPGHRCLNGGTCVDGVNTYNCQCPPEWTGQFCTEDVDECQLQ 281
Query 87 GAICSGNATCDQE--ALSCVCKPGYTGEDCTELSPECSGLCTVGGSVIRDSVS 137
C TC SCVC G+TGE C++ +C+ G+ D V+
Sbjct 282 PNACHNGGTCFNTLGGHSCVCVNGWTGESCSQNIDDCATAVCFHGATCHDRVA 334
Score = 33.9 bits (76), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 38/107 (35%), Positives = 44/107 (41%), Gaps = 8/107 (7%)
Query 30 SNTCECTPEFPAANC---VDLCSGVECSNDGKC--DPATGKCTCLAGWGGPQCIVPQP-C 83
S +C C P F C VD C C G C A+ CTC G+GG C P C
Sbjct 870 SFSCSCLPGFAGPRCARDVDECLSNPC-GPGTCTDHVASFTCTCPPGYGGFHCEQDLPDC 928
Query 84 GTEGAICSGNATCDQEALSCVCKPGYTGEDCT-ELSPECSGLCTVGG 129
G + SC+C+PGYTG C E P S C GG
Sbjct 929 SPSSCFNGGTCVDGVNSFSCLCRPGYTGAHCQHEADPCLSRPCLHGG 975
Score = 32.7 bits (73), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 38/96 (39%), Gaps = 13/96 (13%)
Query 32 TCECTPEFPAANC---VDLCSGVECSNDGKCDPATGK--CTCLAGWGGPQCIV------P 80
CEC P + NC VD C+ C + G C + C+C G G C + P
Sbjct 1107 MCECLPGYNGDNCEDDVDECASQPCQHGGSCIDLVARYLCSCPPGTLGVLCEINEDDCGP 1166
Query 81 QPCGTEGAICSGNATCDQEA--LSCVCKPGYTGEDC 114
P G C N TC C C PGYTG C
Sbjct 1167 GPPLDSGPRCLHNGTCVDLVGGFRCTCPPGYTGLRC 1202
Score = 31.6 bits (70), Expect = 0.71, Method: Compositional matrix adjust.
Identities = 44/136 (32%), Positives = 56/136 (41%), Gaps = 12/136 (8%)
Query 11 ANCCESDSDCNAPNGVCNSSNTCECTPEFPAANC---VDLCS--GVECSNDGKCDPATGK 65
+N C D+ C+ N V N C C P F C VD CS C + G+C G
Sbjct 357 SNPCHEDAICDT-NPV-NGRAICTCPPGFTGGACDQDVDECSIGANPCEHLGRCVNTQGS 414
Query 66 --CTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQE--ALSCVCKPGYTGEDCTELSPEC 121
C C G+ GP+C G C ATC +C+C G+TG C EC
Sbjct 415 FLCQCGRGYTGPRCETDVNECLSGP-CRNQATCLDRIGQFTCICMAGFTGTYCEVDIDEC 473
Query 122 SGLCTVGGSVIRDSVS 137
V G V +D V+
Sbjct 474 QSSPCVNGGVCKDRVN 489
Score = 31.2 bits (69), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 38/113 (33%), Positives = 45/113 (39%), Gaps = 18/113 (15%)
Query 32 TCECTPEFPAANC---VDLCSGVECSNDGKC--DPATGKCTCLAGWGGPQCIV------P 80
+C C F + C VD C+ C N KC P +C C G+ G C P
Sbjct 492 SCTCPSGFSGSTCQLDVDECASTPCRNGAKCVDQPDGYECRCAEGFEGTLCDRNVDDCSP 551
Query 81 QPCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTELSPEC-SGLCTVGGSVI 132
PC G G A+ SC C PGYTG C EC S C GG +
Sbjct 552 DPC-HHGRCVDGIAS-----FSCACAPGYTGTRCESQVDECRSQPCRHGGKCL 598
Score = 31.2 bits (69), Expect = 0.93, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 27/64 (42%), Gaps = 5/64 (7%)
Query 62 ATGKCTCLAGWGGPQCIVPQPCGTEGAICSGNATCD---QEALSCVCKPGYTGEDCTELS 118
A C C G+ GP C +P PC + + C+ A C C C PGY G C
Sbjct 102 ARFSCRCPRGFRGPDCSLPDPCLS--SPCAHGARCSVGPDGRFLCSCPPGYQGRSCRSDV 159
Query 119 PECS 122
EC
Sbjct 160 DECR 163
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 31/101 (30%), Positives = 41/101 (40%), Gaps = 16/101 (15%)
Query 29 SSNTCECTPEFPAANCVDLCSGVECSNDGKCDPA----TGKCTCLAGWGGPQCIVP--QP 82
S +C P P C+ C + G C PA +C C GW GP+C P P
Sbjct 1320 SGPSCRSFPGSPPGASNASCAAAPCLHGGSCRPAPLAPFFRCACAQGWTGPRCEAPAAAP 1379
Query 83 CGTEGAIC--------SGNATCDQEALSCVCKPGYTGEDCT 115
+E C G+ CD+E S C G+ G DC+
Sbjct 1380 EVSEEPRCPRAACQAKRGDQRCDRECNSPGC--GWDGGDCS 1418
Score = 28.1 bits (61), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 40/116 (34%), Positives = 48/116 (41%), Gaps = 18/116 (15%)
Query 29 SSNTCECTPEFPAANC---VDLCSGVECSNDGKCDPATGK--CTCLAGWGGPQCIV---- 79
+S +C C P + C VD C C + GKC K C C +G G C V
Sbjct 564 ASFSCACAPGYTGTRCESQVDECRSQPCRHGGKCLDLVDKYLCRCPSGTTGVNCEVNIDD 623
Query 80 --PQPCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTELSPEC-SGLCTVGGSVI 132
PC T G G D CVC+PG+TG C EC S C GGS +
Sbjct 624 CASNPC-TFGVCRDGINRYD-----CVCQPGFTGPLCNVEINECASSPCGEGGSCV 673
> 7290390
Length=2634
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 43/128 (33%), Positives = 56/128 (43%), Gaps = 24/128 (18%)
Query 2 SGSNCETH----EANCCESDSDCNAPNGVCNSSNTCECTPEFPAANC---VDLCSGVECS 54
+G CET+ E++ C+++ C G C C P F C +D C C
Sbjct 481 TGPRCETNINECESHPCQNEGSCLDDPGTFR----CVCMPGFTGTQCEIDIDECQSNPCL 536
Query 55 NDGKC-DPATG-KCTCLAGWGGPQCIV------PQPCGTEGAICSGNATCDQEALSCVCK 106
NDG C D G KC+C G+ G +C + QPC G IC + SC C
Sbjct 537 NDGTCHDKINGFKCSCALGFTGARCQINIDDCQSQPCRNRG-ICHDSIA----GYSCECP 591
Query 107 PGYTGEDC 114
PGYTG C
Sbjct 592 PGYTGTSC 599
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 47/143 (32%), Positives = 62/143 (43%), Gaps = 26/143 (18%)
Query 22 APNGVC---NSSNTCECTPEFPAANC---VDLCSGVECSNDGKCDPATGK--CTCLAGWG 73
A NGVC + CEC F A+C VD C+ C N+G+C+ + C C G+
Sbjct 725 ANNGVCIDQVNGYKCECPRGFYDAHCLSDVDECASNPCVNEGRCEDGINEFICHCPPGYT 784
Query 74 GPQCIV------PQPCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTELSPEC-SGLCT 126
G +C + PC G C A SC C PGYTG+ C +C + C
Sbjct 785 GKRCELDIDECSSNPC-QHGGTCYDKLN----AFSCQCMPGYTGQKCETNIDDCVTNPCG 839
Query 127 VGGSVIRDSVSECKVDCYSACCK 149
GG+ I KV+ Y CK
Sbjct 840 NGGTCID------KVNGYKCVCK 856
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 37/124 (29%), Positives = 52/124 (41%), Gaps = 19/124 (15%)
Query 11 ANCCESDSDCNAPNGVCNSSNTCECTPEFPAANC---VDLC-SGVECSNDGKC--DPATG 64
+N C +D+ C+ N S C C + +C +D C G C ++G C P +
Sbjct 415 SNPCHADAICDT--SPINGSYACSCATGYKGVDCSEDIDECDQGSPCEHNGICVNTPGSY 472
Query 65 KCTCLAGWGGPQCIV------PQPCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTELS 118
+C C G+ GP+C PC EG+ D CVC PG+TG C
Sbjct 473 RCNCSQGFTGPRCETNINECESHPCQNEGSCLD-----DPGTFRCVCMPGFTGTQCEIDI 527
Query 119 PECS 122
EC
Sbjct 528 DECQ 531
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 47/152 (30%), Positives = 66/152 (43%), Gaps = 23/152 (15%)
Query 2 SGSNCETHE---ANCCESDSDCNAPNGVCNSSNTCECTPEFPAANC---VDLCSGVECSN 55
+G CET ++ C + + C A G +SS TC C P F C ++ C C
Sbjct 171 TGERCETKNLCASSPCRNGATCTALAG--SSSFTCSCPPGFTGDTCSYDIEECQSNPCKY 228
Query 56 DGKCDPATG--KCTCLAGWGGPQCIV------PQPCGTEGAICSGNATCDQEALSCVCKP 107
G C G +C C G+ G C P PC G IC N + C C
Sbjct 229 GGTCVNTHGSYQCMCPTGYTGKDCDTKYKPCSPSPC-QNGGICRSNGL----SYECKCPK 283
Query 108 GYTGEDCTELSPECSG-LCTVGGSVIRDSVSE 138
G+ G++C + +C G LC GG+ I D +S+
Sbjct 284 GFEGKNCEQNYDDCLGHLCQNGGTCI-DGISD 314
Score = 33.5 bits (75), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 35/101 (34%), Positives = 42/101 (41%), Gaps = 12/101 (11%)
Query 32 TCECTPEFPAANC---VDLCSGVECSNDGKC-DPATG-KCTCLAGWGGPQCIVPQ-PCGT 85
+C+C P + C +D C C N G C D G KC C + G C PC +
Sbjct 814 SCQCMPGYTGQKCETNIDDCVTNPCGNGGTCIDKVNGYKCVCKVPFTGRDCESKMDPCAS 873
Query 86 EGAICSGNATCDQEA----LSCVCKPGYTGEDCTELSPECS 122
C A C + SC CK GYTG C E ECS
Sbjct 874 N--RCKNEAKCTPSSNFLDFSCTCKLGYTGRYCDEDIDECS 912
Score = 33.1 bits (74), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 46/153 (30%), Positives = 57/153 (37%), Gaps = 21/153 (13%)
Query 2 SGSNCETHEANCCESDSDCNAPNGVCNSSN---TCECTPEFPAANC---VDLCSGVECSN 55
SG+NC+ ++ N C+S+ N C+ N TC C F C VD C C N
Sbjct 1053 SGANCQ-YKLNKCDSNPCLNG--ATCHEQNNEYTCHCPSGFTGKQCSEYVDWCGQSPCEN 1109
Query 56 DGKCDPATGK--CTCLAGWGGPQCIVPQPCGTEGAICSG--------NATCDQEALS--C 103
C + C C AGW G C V + A G N TC S C
Sbjct 1110 GATCSQMKHQFSCKCSAGWTGKLCDVQTISCQDAADRKGLSLRQLCNNGTCKDYGNSHVC 1169
Query 104 VCKPGYTGEDCTELSPECSGLCTVGGSVIRDSV 136
C GY G C + EC G RD +
Sbjct 1170 YCSQGYAGSYCQKEIDECQSQPCQNGGTCRDLI 1202
Score = 32.3 bits (72), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 31/108 (28%), Positives = 47/108 (43%), Gaps = 10/108 (9%)
Query 33 CECTPEFPAANC---VDLCSGVECSNDGKCDPATG----KCTCLAGWGGPQCIVPQPCGT 85
C C F +C +D C+ C N+ KC P++ CTC G+ G C +
Sbjct 853 CVCKVPFTGRDCESKMDPCASNRCKNEAKCTPSSNFLDFSCTCKLGYTGRYCDEDIDECS 912
Query 86 EGAICSGNATCDQE--ALSCVCKPGYTGEDCTELSPECSGL-CTVGGS 130
+ C A+C + C+C GY G DC + +C+ C GG+
Sbjct 913 LSSPCRNGASCLNVPGSYRCLCTKGYEGRDCAINTDDCASFPCQNGGT 960
Score = 31.6 bits (70), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 43/145 (29%), Positives = 59/145 (40%), Gaps = 20/145 (13%)
Query 2 SGSNCETHEANCCESD-----SDCNAPNGVCNSSNTCECTPEFPAANC---VDLCSGVEC 53
SG NC+T++ +C ES S + NG C C + ANC ++ C C
Sbjct 1015 SGINCQTNDEDCTESSCLNGGSCIDGINGY-----NCSCLAGYSGANCQYKLNKCDSNPC 1069
Query 54 SNDGKCDPATGK--CTCLAGWGGPQCI-VPQPCGTEGAICSGNATCDQ--EALSCVCKPG 108
N C + C C +G+ G QC CG + C ATC Q SC C G
Sbjct 1070 LNGATCHEQNNEYTCHCPSGFTGKQCSEYVDWCGQ--SPCENGATCSQMKHQFSCKCSAG 1127
Query 109 YTGEDCTELSPECSGLCTVGGSVIR 133
+TG+ C + C G +R
Sbjct 1128 WTGKLCDVQTISCQDAADRKGLSLR 1152
Score = 29.3 bits (64), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 32/104 (30%), Positives = 40/104 (38%), Gaps = 16/104 (15%)
Query 29 SSNTCECTPEFPAANC---VDLCSGVECSNDGKCDPATGK--CTCLAGWGGPQCIV---- 79
+S C C P + C ++ C C DG C G C C AG G C V
Sbjct 621 NSFKCLCDPGYTGYICQKQINECESNPCQFDGHCQDRVGSYYCQCQAGTSGKNCEVNVNE 680
Query 80 --PQPCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTELSPEC 121
PC GA C + C C PG+TG+ C + EC
Sbjct 681 CHSNPC-NNGATCIDGIN----SYKCQCVPGFTGQHCEKNVDEC 719
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 36/124 (29%), Positives = 49/124 (39%), Gaps = 12/124 (9%)
Query 25 GVCNS---SNTCECTPEFPAANC---VDLCSGVECSNDGKCDPATGK--CTCLAGWGGPQ 76
G+C S S C+C F NC D C G C N G C C C + G
Sbjct 268 GICRSNGLSYECKCPKGFEGKNCEQNYDDCLGHLCQNGGTCIDGISDYTCRCPPNFTGRF 327
Query 77 CI--VPQPCGTEGAICSGNATCDQE--ALSCVCKPGYTGEDCTELSPECSGLCTVGGSVI 132
C V + + +C ATC + SC+C G+ G DC+ + +C G+
Sbjct 328 CQDDVDECAQRDHPVCQNGATCTNTHGSYSCICVNGWAGLDCSNNTDDCKQAACFYGATC 387
Query 133 RDSV 136
D V
Sbjct 388 IDGV 391
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 38/126 (30%), Positives = 49/126 (38%), Gaps = 24/126 (19%)
Query 24 NGVCN---SSNTCECTPEFPAANC---VDLCSGVECSNDGKCDPATG--KCTCLAGWGGP 75
NG C +S+ C C+ + + C +D C C N G C G +C C G+ G
Sbjct 1157 NGTCKDYGNSHVCYCSQGYAGSYCQKEIDECQSQPCQNGGTCRDLIGAYECQCRQGFQGQ 1216
Query 76 QCIV------PQPCGTEGAICSGNATCDQEAL--SCVCKPGYTGEDCTELSPECS-GLCT 126
C + P PC G TC + SC C PG G C +C G C
Sbjct 1217 NCELNIDDCAPNPCQNGG-------TCHDRVMNFSCSCPPGTMGIICEINKDDCKPGACH 1269
Query 127 VGGSVI 132
GS I
Sbjct 1270 NNGSCI 1275
Score = 28.5 bits (62), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 33/111 (29%), Positives = 41/111 (36%), Gaps = 9/111 (8%)
Query 33 CECTPEFPAANC---VDLCSGVECSNDGKCD----PATGKCTCLAGWGGPQCIVP-QPCG 84
C C P F A C ++ C CSN G D C C G G C C
Sbjct 1283 CVCQPGFVGARCEGDINECLSNPCSNAGTLDCVQLVNNYHCNCRPGHMGRHCEHKVDFCA 1342
Query 85 TEGAICSGNATCDQEALSCVCKPGYTGEDCTELSPEC-SGLCTVGGSVIRD 134
GN Q C+C G+ G++C +C S C VG V+ D
Sbjct 1343 QSPCQNGGNCNIRQSGHHCICNNGFYGKNCELSGQDCDSNPCRVGNCVVAD 1393
> CE29452_1
Length=650
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 34/104 (32%), Positives = 47/104 (45%), Gaps = 9/104 (8%)
Query 16 SDSDCNAPNGVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGP 75
S C NG C +C+C + +NC D + C + GKC + G C C +GW G
Sbjct 531 SQQACQCVNGDCLDDGSCQCWKGWRGSNCTDKKCAIGCEDRGKC-ASDGSCKCSSGWNGE 589
Query 76 QCIVPQPCGTEGAICSGNATCDQEA----LSCVCKPGYTGEDCT 115
C + C + CSG C + SC C+ G TG DC+
Sbjct 590 NCAI-DGCPNQ---CSGKGECGMDRRSSEWSCRCQAGSTGVDCS 629
Score = 35.8 bits (81), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 29/82 (35%), Positives = 39/82 (47%), Gaps = 4/82 (4%)
Query 33 CECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSG 92
C C P F C + V CS +G + G C C +G+ G +C + E A C+G
Sbjct 332 CHCAPGFTGRTCDEAVCPVVCSGNGVF--SGGICVCKSGFKGKECEMRHNW-CEVADCNG 388
Query 93 NATCDQEALSCVCKPGYTGEDC 114
CD + C C PG+TGE C
Sbjct 389 RGRCDTDG-RCRCNPGWTGEAC 409
Score = 32.7 bits (73), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 34/79 (43%), Gaps = 6/79 (7%)
Query 2 SGSNCETHEANCCESDSDCNAPNGVCNSSNTCECTPEFPAANC-VDLCSGVECSNDGKCD 60
G CE N CE +DCN G C++ C C P + C + C C + G C
Sbjct 370 KGKECEMRH-NWCEV-ADCNG-RGRCDTDGRCRCNPGWTGEACELRACPHASCHDRGVC- 425
Query 61 PATGKCTCLAGWGGPQCIV 79
G C C+ GW G C V
Sbjct 426 -VNGTCYCMDGWRGNDCSV 443
> 7292090
Length=601
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 32/91 (35%), Positives = 43/91 (47%), Gaps = 16/91 (17%)
Query 45 VDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSG---NATCDQEAL 101
+ + C NDG+ AT C C G+ G +C + + C + TC L
Sbjct 322 IKVLEDERCMNDGEYRAATDLCICPTGFKGSRCEIRE--------CHNYCVHGTCQMSEL 373
Query 102 S---CVCKPGYTGEDCTELSPECSGLCTVGG 129
+ C C+PG+ GE C ELS CSGLC GG
Sbjct 374 AYPKCYCQPGFKGERC-ELSV-CSGLCLNGG 402
> 7296540_1
Length=593
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 42/89 (47%), Gaps = 4/89 (4%)
Query 30 SNTCECTPEFPAANCVDL-CSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTE-G 87
S C+C + C ++ C CS G C A G C C GW GP C +
Sbjct 375 SGKCQCMRGYKGKFCEEVDCPHPNCSGHGFC--ADGTCICKKGWKGPDCATMDQDALQCL 432
Query 88 AICSGNATCDQEALSCVCKPGYTGEDCTE 116
CSG+ T D + +C C+ ++G+DC++
Sbjct 433 PDCSGHGTFDLDTQTCTCEAKWSGDDCSK 461
Score = 36.6 bits (83), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 33/94 (35%), Positives = 44/94 (46%), Gaps = 8/94 (8%)
Query 33 CECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSG 92
C+C P F +C + V CS G + G+C C GW G +C + E A CSG
Sbjct 313 CQCNPGFGGDDCSESVCPVLCSQHG--EYTNGECICNPGWKGKECSLRHD-ECEVADCSG 369
Query 93 NATCDQEALSCVCKPGYTGEDCTEL---SPECSG 123
+ C + C C GY G+ C E+ P CSG
Sbjct 370 HGHC--VSGKCQCMRGYKGKFCEEVDCPHPNCSG 401
Score = 36.2 bits (82), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 35/108 (32%), Positives = 46/108 (42%), Gaps = 23/108 (21%)
Query 33 CECTPEFPAANCV---DLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIV---PQPCGTE 86
C C P + C D C +CS G C +GKC C+ G+ G C P P
Sbjct 344 CICNPGWKGKECSLRHDECEVADCSGHGHC--VSGKCQCMRGYKGKFCEEVDCPHP---- 397
Query 87 GAICSGNATCDQEALSCVCKPGYTGEDCTELS-------PECSGLCTV 127
CSG+ C +C+CK G+ G DC + P+CSG T
Sbjct 398 --NCSGHGFCADG--TCICKKGWKGPDCATMDQDALQCLPDCSGHGTF 441
Score = 31.2 bits (69), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 33/72 (45%), Gaps = 14/72 (19%)
Query 53 CSNDGKCDPATGKCTCLAGWGGPQC---IVPQPCGTEGAICSGNATCDQEALSCVCKPGY 109
CS +G+C G C C G+GG C + P C G +G C+C PG+
Sbjct 302 CSGNGQC--LLGHCQCNPGFGGDDCSESVCPVLCSQHGEYTNG---------ECICNPGW 350
Query 110 TGEDCTELSPEC 121
G++C+ EC
Sbjct 351 KGKECSLRHDEC 362
> CE28688
Length=2192
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 42/143 (29%), Positives = 59/143 (41%), Gaps = 15/143 (10%)
Query 14 CESDSDCNAPNGVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPA---TGKCTCLA 70
C ++S C NG + C+C P F C ECSN+ KC T C C
Sbjct 1922 CTNNSKCTITNG---THFECDCKPGFKGLRCEQETKCSECSNEAKCIKKPSGTVICQCPQ 1978
Query 71 GWGGPQC--IVPQPCG----TEGAIC-SGNATCDQEALSCVCKPGYTGEDCTELSPECSG 123
G GG C I C G C + + ++ + +C+C PG+ G C+ S C
Sbjct 1979 GLGGEYCEKITATSCHQLECKNGGYCLKPDPSANRTSPTCLCSPGFKGLLCS--SNACEN 2036
Query 124 LCTVGGSVIRDSVSECKVDCYSA 146
C G+ D E + +CY A
Sbjct 2037 FCLHDGNCTLDEYFEPQCECYQA 2059
Score = 36.6 bits (83), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 34/70 (48%), Gaps = 9/70 (12%)
Query 53 CSNDGKC--DPAT-GKCTCLAGWGGPQCIVPQPCGTEGAICSGNATC---DQEALSCVCK 106
C N G+C P GKC CL + GP+C VP C C+ N+ C + C CK
Sbjct 1885 CHNGGRCLDTPGYPGKCKCLPRFAGPRCDVPVQCDD---YCTNNSKCTITNGTHFECDCK 1941
Query 107 PGYTGEDCTE 116
PG+ G C +
Sbjct 1942 PGFKGLRCEQ 1951
Score = 28.5 bits (62), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 31/101 (30%), Positives = 44/101 (43%), Gaps = 13/101 (12%)
Query 27 CNSSNTCECTPEFPAANC-----VDLCSGVECSNDGKCD---PATGKCTCLAGWGGPQCI 78
C S + CEC + +C +C G C + G CD P +C+C G G +C
Sbjct 1739 CRSYSFCECDKGWTGPHCRHKADAKVCYG-HCFSGGACDGEGPLNLRCSCGDGLTGNRCQ 1797
Query 79 --VPQPCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTEL 117
V C G CS A ++ C+C G+TG+ C E
Sbjct 1798 NCVGHEC-LNGGFCS-YANSNRSLPHCICPSGFTGDHCEEY 1836
> HsM6912258
Length=1198
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 19/35 (54%), Positives = 22/35 (62%), Gaps = 3/35 (8%)
Query 50 GVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCG 84
G C N G+C+P TG+C C AGW G QC Q CG
Sbjct 28 GPHCVNGGRCNPGTGQCVCPAGWVGEQC---QHCG 59
> Hs21450848
Length=1198
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 19/35 (54%), Positives = 22/35 (62%), Gaps = 3/35 (8%)
Query 50 GVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCG 84
G C N G+C+P TG+C C AGW G QC Q CG
Sbjct 28 GPHCVNGGRCNPGTGQCVCPAGWVGEQC---QHCG 59
> Hs20539391
Length=849
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 37/125 (29%), Positives = 49/125 (39%), Gaps = 14/125 (11%)
Query 27 CNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQ---PC 83
C + N C+C P + +NC +C N GKC C CL G GG C PC
Sbjct 567 CVAPNICKCKPGYIGSNCQTALCDPDCKNHGKCI-KPNICQCLPGHGGATCDEEHCNPPC 625
Query 84 GTEGAICSGNATCDQEALSCVCKPGYTGEDCTELSPECSGLCTVGGSVIRDSVSECKVDC 143
G +GN C C G+ G C + C+ C GG + + +CK
Sbjct 626 QHGGTCLAGNL--------CTCPYGFVGPRCETMV--CNRHCENGGQCLTPDICQCKPGW 675
Query 144 YSACC 148
Y C
Sbjct 676 YGPTC 680
Score = 31.2 bits (69), Expect = 0.85, Method: Compositional matrix adjust.
Identities = 36/117 (30%), Positives = 46/117 (39%), Gaps = 12/117 (10%)
Query 25 GVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGP--QCIVPQP 82
G CN NTC C F +C + C N G C C C G+ G Q + P
Sbjct 693 GSCNKPNTCLCPNGFFGEHCQNAFCHPPCKNGGHCM-RNNVCVCREGYTGRRFQKSICDP 751
Query 83 CGTEGAICSGNATCDQEALSCVCKPGYTGEDCTELSPECSGLCTVGGSVIRDSVSEC 139
G C G +TC C G++G+ C +P C C GG I S+ C
Sbjct 752 TCMNGGKCVGPSTCS-------CPSGWSGKRCN--TPICLQKCKNGGECIAPSICHC 799
Score = 28.9 bits (63), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 37/133 (27%), Positives = 51/133 (38%), Gaps = 16/133 (12%)
Query 25 GVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKC-DPATGKCTCLAGWGGPQCIVP--- 80
G C +N C C + C N GKC P+T C+C +GW G +C P
Sbjct 725 GHCMRNNVCVCREGYTGRRFQKSICDPTCMNGGKCVGPST--CSCPSGWSGKRCNTPICL 782
Query 81 QPCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTELSPECSGLCTVGGSVIRDSVSECK 140
Q C G C + C C + G C P C+ C GG I +V C+
Sbjct 783 QKC-KNGGECIAPSICH-------CPSSWEGVRCQ--IPICNPKCLYGGRCIFPNVCSCR 832
Query 141 VDCYSACCKHLIE 153
+ C+ I+
Sbjct 833 TEYSGVKCEKKIQ 845
> Hs6005950
Length=379
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 33/95 (34%), Positives = 40/95 (42%), Gaps = 12/95 (12%)
Query 25 GVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIV---PQ 81
G+C + C C P F NC C N G C GKC C G G QC + PQ
Sbjct 222 GLCVTPGFCICPPGFYGVNCDKANCSTTCFNGGTCF-YPGKCICPPGLEGEQCEISKCPQ 280
Query 82 PCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTE 116
PC G C G + C C GY G+ C++
Sbjct 281 PC-RNGGKCIGKSKCK-------CSKGYQGDLCSK 307
Score = 30.8 bits (68), Expect = 0.97, Method: Compositional matrix adjust.
Identities = 39/115 (33%), Positives = 45/115 (39%), Gaps = 8/115 (6%)
Query 25 GVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCG 84
G CN CEC F +C C N G C G C C G+ G C C
Sbjct 190 GFCNERRICECPDGFHGPHCEKALCTPRCMNGGLCV-TPGFCICPPGFYGVNCDKAN-CS 247
Query 85 TEGAICSGNATCDQEALSCVCKPGYTGEDCTELSPECSGLCTVGGSVIRDSVSEC 139
T C TC C+C PG GE C E+S +C C GG I S +C
Sbjct 248 T---TCFNGGTCFYPG-KCICPPGLEGEQC-EIS-KCPQPCRNGGKCIGKSKCKC 296
> Hs16158959
Length=143
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 35/113 (30%), Positives = 49/113 (43%), Gaps = 13/113 (11%)
Query 11 ANCCESDSDCNAPNG----VCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKC 66
C E S+C+ P G +CN + C + A C C VE N +C G C
Sbjct 32 GKCQEGKSECSCPAGCRVILCNEN----CLEDAYGAGCTSECQCVE-ENTLECSAKNGSC 86
Query 67 TCLAGWGGPQCIVPQPCGTEGAI----CSGNATCDQEALSCVCKPGYTGEDCT 115
TC +G+ G +C G EG C C+++ +C C P YT + CT
Sbjct 87 TCKSGYQGNRCQKDGLWGPEGWFSSAPCENGGQCNKKTGNCDCTPDYTRKSCT 139
> CE00237
Length=1295
Score = 37.0 bits (84), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 31/103 (30%), Positives = 42/103 (40%), Gaps = 20/103 (19%)
Query 32 TCECTPEFPAANC---VDLCSGVECSNDGKCDPATGK---CTCLAGWGGPQCIVPQPCGT 85
TC C P + C +D+C C NDG C + + C C G+ G +C + P G
Sbjct 256 TCTCKPGYAGKYCEEAIDMCKDYVCQNDGYCAHDSNQMPICYCEQGFTGQRCEIECPSGF 315
Query 86 EGAIC----------SGNATCDQEAL----SCVCKPGYTGEDC 114
G C N TC + CVC+P Y G+ C
Sbjct 316 GGIHCDLPLQRPHCSRSNGTCYNDGRCINGFCVCEPDYIGDRC 358
Score = 33.5 bits (75), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 31/101 (30%), Positives = 42/101 (41%), Gaps = 19/101 (18%)
Query 28 NSSNTCECTPEFPAANCVD--LCSGVECSNDGKCDPATG--KCTCLAGWGGPQCIVP--- 80
NS +C C F NC LC+ C+N G C+ G +C C G+ G C +
Sbjct 389 NSGYSCHCPLGFYGLNCEQHLLCTPTTCANGGTCEGVNGVIRCNCPNGFSGDYCEIKDRQ 448
Query 81 ----QPCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTEL 117
PC G +C C+ C+ GYTG C E+
Sbjct 449 LCSRHPC-KNGGVCKNTGYCE-------CQYGYTGPTCEEV 481
Score = 28.5 bits (62), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 30/69 (43%), Gaps = 7/69 (10%)
Query 55 NDGKCDP-----ATGKCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQEA--LSCVCKP 107
N G+C P +C C +G+ G C + C N+TC +C CKP
Sbjct 202 NHGRCIPNRDEDKNFRCVCDSGYEGEFCNKDKNECLIEETCVNNSTCFNLHGDFTCTCKP 261
Query 108 GYTGEDCTE 116
GY G+ C E
Sbjct 262 GYAGKYCEE 270
> HsM4503339
Length=383
Score = 37.0 bits (84), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 40/129 (31%), Positives = 56/129 (43%), Gaps = 19/129 (14%)
Query 24 NGVCNSSNTCECTPEFPAANC---VDLCSGVECSNDGKC---DPATGKCTCLAGWGGPQC 77
+G+C C CT + C V CS C+N+G C D +C+C G+ G C
Sbjct 65 HGLCGEPGQCICTDGWDGELCDRDVRACSSAPCANNGTCVSLDGGLYECSCAPGYSGKDC 124
Query 78 IVPQ-PCGTEGAICSGNATC-DQEAL----SCVCKPGYTGEDCTELSPECS-------GL 124
PC G+ C TC D E SC+C PG++G C ++ C+ G+
Sbjct 125 QKKDGPCVINGSPCQHGGTCVDDEGRASHASCLCPPGFSGNFCEIVANSCTPNPCENDGV 184
Query 125 CTVGGSVIR 133
CT G R
Sbjct 185 CTDIGGDFR 193
Score = 35.8 bits (81), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 39/115 (33%), Positives = 48/115 (41%), Gaps = 12/115 (10%)
Query 20 CNAPNGVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIV 79
CN NG C N C C + C D C G C G+C C GW G C
Sbjct 30 CNPQNGFCEDDNVCRCHVGWQGPLC-DQCVTSPGCLHGLCG-EPGQCICTDGWDGELCDR 87
Query 80 P-QPCGTEGAICSGNATC---DQEALSCVCKPGYTGEDCTELSPECSGLCTVGGS 130
+ C + A C+ N TC D C C PGY+G+DC + G C + GS
Sbjct 88 DVRACSS--APCANNGTCVSLDGGLYECSCAPGYSGKDCQKK----DGPCVINGS 136
Score = 31.2 bits (69), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 41/136 (30%), Positives = 56/136 (41%), Gaps = 19/136 (13%)
Query 2 SGSNCETHEANCCESDSDCNAPNGVC------NSSNTCECTPEFPAANC---VDLCSGVE 52
SG +C+ + C + S C G C S +C C P F C + C+
Sbjct 120 SGKDCQKKDGPCVINGSPCQH-GGTCVDDEGRASHASCLCPPGFSGNFCEIVANSCTPNP 178
Query 53 CSNDGKCDPATG--KCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQE---ALSCVCKP 107
C NDG C G +C C AG+ C P + C TC Q + C+CKP
Sbjct 179 CENDGVCTDIGGDFRCRCPAGFIDKTCSRPV-TNCASSPCQNGGTCLQHTQVSYECLCKP 237
Query 108 GYTGEDCTE---LSPE 120
+TG C + LSP+
Sbjct 238 EFTGLTCVKKRALSPQ 253
> Hs13249344
Length=2471
Score = 37.0 bits (84), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 48/146 (32%), Positives = 62/146 (42%), Gaps = 25/146 (17%)
Query 2 SGSNCETHEANC----CESDSDCN-APNGVCNSSNTCECTPEFPAANC---VDLCSGVEC 53
+G NC+T A C CE+ + C +PN S TC C P + C +D C C
Sbjct 826 TGKNCQTVLAPCSPNPCENAAVCKESPNF---ESYTCLCAPGWQGQRCTIDIDECISKPC 882
Query 54 SNDGKCDPATG--KCTCLAGWGGPQCI------VPQPCGTEGAICSGNATCDQEALSCVC 105
N G C G C C G+ G C + PC G+ G T SC+C
Sbjct 883 MNHGLCHNTQGSYMCECPPGFSGMDCEEDIDDCLANPCQNGGSCMDGVNT-----FSCLC 937
Query 106 KPGYTGEDCTELSPEC-SGLCTVGGS 130
PG+TG+ C EC S C GG+
Sbjct 938 LPGFTGDKCQTDMNECLSEPCKNGGT 963
Score = 33.5 bits (75), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 36/99 (36%), Positives = 44/99 (44%), Gaps = 7/99 (7%)
Query 30 SNTCECTPEFPAANC---VDLCSGVECSNDGKC-DPATG-KCTCLAGWGGPQC-IVPQPC 83
S TC C P + A C +D C C NDG+C D G +C C G G C I C
Sbjct 590 SYTCICNPGYMGAICSDQIDECYSSPCLNDGRCIDLVNGYQCNCQPGTSGVNCEINFDDC 649
Query 84 GTEGAICSGNATCDQEALSCVCKPGYTGEDCTELSPECS 122
+ I G SCVC PG+TG+ C EC+
Sbjct 650 ASNPCI-HGICMDGINRYSCVCSPGFTGQRCNIDIDECA 687
Score = 32.7 bits (73), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 32/69 (46%), Gaps = 9/69 (13%)
Query 53 CSNDGKC---DPATGKCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQEAL----SCVC 105
C N+G C TG C C G+ G C PC E C TC +A+ +C C
Sbjct 35 CVNEGMCVTYHNGTGYCKCPEGFLGEYCQHRDPC--EKNRCQNGGTCVAQAMLGKATCRC 92
Query 106 KPGYTGEDC 114
G+TGEDC
Sbjct 93 ASGFTGEDC 101
Score = 32.3 bits (72), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 41/97 (42%), Gaps = 12/97 (12%)
Query 43 NCVDLCSGVECSNDGKCDPATG--KCTCLAGWGGPQCI------VPQPCGTEGAICSGNA 94
N D G C N G+C G C CL G+ G +C + PC +EG++
Sbjct 1225 NIDDCARGPHCLNGGQCMDRIGGYSCRCLPGFAGERCEGDINECLSNPCSSEGSLDCIQL 1284
Query 95 TCDQEALSCVCKPGYTGEDCTELSPECSGL-CTVGGS 130
T D CVC+ +TG C C + C GG+
Sbjct 1285 TND---YLCVCRSAFTGRHCETFVDVCPQMPCLNGGT 1318
Score = 32.3 bits (72), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 34/113 (30%), Positives = 44/113 (38%), Gaps = 8/113 (7%)
Query 33 CECTPEFPAANC---VDLCSGVECSNDGKCDPA--TGKCTCLAGWGGPQCIVP-QPCGTE 86
C C P F + C +D C C N G C T C C W G C C +
Sbjct 246 CNCLPGFEGSTCERNIDDCPNHRCQNGGVCVDGVNTYNCRCPPQWTGQFCTEDVDECLLQ 305
Query 87 GAICSGNATCDQE--ALSCVCKPGYTGEDCTELSPECSGLCTVGGSVIRDSVS 137
C TC CVC G++G+DC+E +C+ GS D V+
Sbjct 306 PNACQNGGTCANRNGGYGCVCVNGWSGDDCSENIDDCAFASCTPGSTCIDRVA 358
Score = 32.0 bits (71), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 33/106 (31%), Positives = 47/106 (44%), Gaps = 13/106 (12%)
Query 28 NSSNTCECTPEFPAANC---VDLCSGVE---CSNDGKCDPATG--KCTCLAGWGGPQC-I 78
N C C + A+C VD C+ C + GKC G C CL G+ GP+C +
Sbjct 396 NGQYICTCPQGYKGADCTEDVDECAMANSNPCEHAGKCVNTDGAFHCECLKGYAGPRCEM 455
Query 79 VPQPCGTEGAICSGNATCDQE--ALSCVCKPGYTGEDCTELSPECS 122
C ++ C +ATC + +C+C PG+ G C EC
Sbjct 456 DINECHSDP--CQNDATCLDKIGGFTCLCMPGFKGVHCELEINECQ 499
Score = 31.2 bits (69), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 32/105 (30%), Positives = 43/105 (40%), Gaps = 16/105 (15%)
Query 29 SSNTCECTPEFPAANC---VDLCSGVECSNDGKCDPATGKCTCL--AGWGGPQCI----- 78
+S TC+C F +C ++ C+ C N G C +CL G+ G C+
Sbjct 969 NSYTCKCQAGFDGVHCENNINECTESSCFNGGTCVDGINSFSCLCPVGFTGSFCLHEINE 1028
Query 79 -VPQPCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTELSPECS 122
PC EG G T C C GYTG++C L CS
Sbjct 1029 CSSHPCLNEGTCVDGLGT-----YRCSCPLGYTGKNCQTLVNLCS 1068
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 30/89 (33%), Positives = 33/89 (37%), Gaps = 7/89 (7%)
Query 33 CECTPEFPAANC---VDLCSGVECSNDGKCDPATG--KCTCLAGWGGPQCIVPQPCGTEG 87
CEC P + NC VD C C N G C KC+C G G C G
Sbjct 1173 CECVPGYQGVNCEYEVDECQNQPCQNGGTCIDLVNHFKCSCPPGTRGLLCEENIDDCARG 1232
Query 88 AICSGNATCDQE--ALSCVCKPGYTGEDC 114
C C SC C PG+ GE C
Sbjct 1233 PHCLNGGQCMDRIGGYSCRCLPGFAGERC 1261
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 30/115 (26%), Positives = 44/115 (38%), Gaps = 10/115 (8%)
Query 6 CETHEANCCESDSDCNAPNGVCNSSNTCECTPEFPAANC---VDLCSGVECSNDGKCDPA 62
C +N CE C +G + CEC + C ++ C C ND C
Sbjct 419 CAMANSNPCEHAGKCVNTDGAFH----CECLKGYAGPRCEMDINECHSDPCQNDATCLDK 474
Query 63 TG--KCTCLAGWGGPQC-IVPQPCGTEGAICSGNATCDQEALSCVCKPGYTGEDC 114
G C C+ G+ G C + C + + +G C+C PG+TG C
Sbjct 475 IGGFTCLCMPGFKGVHCELEINECQSNPCVNNGQCVDKVNRFQCLCPPGFTGPVC 529
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 29/96 (30%), Positives = 39/96 (40%), Gaps = 16/96 (16%)
Query 32 TCECTPEFPAANC---VDLCSGVECSNDGKCDPATGKCTCL--AGWGGPQCIV------P 80
TC C P F +C ++ C C N+G+C + CL G+ GP C +
Sbjct 479 TCLCMPGFKGVHCELEINECQSNPCVNNGQCVDKVNRFQCLCPPGFTGPVCQIDIDDCSS 538
Query 81 QPCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTE 116
PC GA C + C C G+TG C E
Sbjct 539 TPC-LNGAKCIDHPN----GYECQCATGFTGVLCEE 569
Score = 27.7 bits (60), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 29/85 (34%), Positives = 38/85 (44%), Gaps = 9/85 (10%)
Query 53 CSNDGKCDPA---TGKCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQEA--LSCVCKP 107
C N G C T +CTC G+ G +C C + C+ +TC A SC C
Sbjct 115 CLNGGTCHMLSRDTYECTCQVGFTGKECQWTDACLSHP--CANGSTCTTVANQFSCKCLT 172
Query 108 GYTGEDCTELSPECS--GLCTVGGS 130
G+TG+ C EC G C GG+
Sbjct 173 GFTGQKCETDVNECDIPGHCQHGGT 197
Score = 27.7 bits (60), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 45/166 (27%), Positives = 53/166 (31%), Gaps = 56/166 (33%)
Query 33 CECTPEFPAANC---VDLCSGVECSNDGKC--DPATGKCTCLAGWGGPQCIVPQ------ 81
C C + NC V+LCS C N G C A +C C +GW G C VP
Sbjct 1049 CSCPLGYTGKNCQTLVNLCSRSPCKNKGTCVQKKAESQCLCPSGWAGAYCDVPNVSCDIA 1108
Query 82 -----------------------------PCGTEGAIC------------SGNATCDQE- 99
P G G+ C ATC
Sbjct 1109 ASRRGVLVEHLCQHSGVCINAGNTHYCQCPLGYTGSYCEEQLDECASNPCQHGATCSDFI 1168
Query 100 -ALSCVCKPGYTGEDCTELSPECSGL-CTVGGSVIRDSVSECKVDC 143
C C PGY G +C EC C GG+ I D V+ K C
Sbjct 1169 GGYRCECVPGYQGVNCEYEVDECQNQPCQNGGTCI-DLVNHFKCSC 1213
Score = 27.7 bits (60), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 29/65 (44%), Gaps = 6/65 (9%)
Query 62 ATGKCTCLAGWGGPQCIVPQPCGT----EGAICSGNATCDQEALSCVCKPGYTGEDCTEL 117
A+ C C G G C + C + +GA+C N Q C C GY G DCTE
Sbjct 358 ASFSCMCPEGKAGLLCHLDDACISNPCHKGALCDTNPLNGQ--YICTCPQGYKGADCTED 415
Query 118 SPECS 122
EC+
Sbjct 416 VDECA 420
Score = 27.7 bits (60), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 37/102 (36%), Gaps = 22/102 (21%)
Query 21 NAPNGVCNSSNTCECTPEFPAANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVP 80
N P+G C C P F A C C V+C + C+ GP+C P
Sbjct 1324 NMPDGF-----ICRCPPGFSGARCQSSCGQVKCRKGEQ---------CVHTASGPRCFCP 1369
Query 81 QP----CGTEGAICSGNATCDQEA----LSCVCKPGYTGEDC 114
P G + C +C + SC C P ++G C
Sbjct 1370 SPRDCESGCASSPCQHGGSCHPQRQPPYYSCQCAPPFSGSRC 1411
> Hs8393264
Length=618
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 39/122 (31%), Positives = 47/122 (38%), Gaps = 12/122 (9%)
Query 2 SGSNCETHEANCCESDSDCNAPNGVC---NSSNTCECTPEFPAANC---VDLCSGVECSN 55
GSNCE C S C G+C + C C F C +D C+G C+N
Sbjct 346 QGSNCEKRVDRC--SLQPCRN-GGLCLDLGHALRCRCRAGFAGPRCEHDLDDCAGRACAN 402
Query 56 DGKCDPATG--KCTCLAGWGGPQCI-VPQPCGTEGAICSGNATCDQEALSCVCKPGYTGE 112
G C G +C+C G+GG C PC G L C C PGY G
Sbjct 403 GGTCVEGGGAHRCSCALGFGGRDCRERADPCAARPCAHGGRCYAHFSGLVCACAPGYMGA 462
Query 113 DC 114
C
Sbjct 463 RC 464
Score = 36.2 bits (82), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 36/109 (33%), Positives = 47/109 (43%), Gaps = 22/109 (20%)
Query 28 NSSNTCECTPEFPAANC---VDLCSGVECSNDGKCDPA--TGKCTCLAGWGGPQCIVPQP 82
+S+ C C P F +NC VD CS C N G C +C C AG+ GP+C
Sbjct 334 DSAYICHCPPGFQGSNCEKRVDRCSLQPCRNGGLCLDLGHALRCRCRAGFAGPRC----- 388
Query 83 CGTE-------GAICSGNATCDQE--ALSCVCKPGYTGEDCTELSPECS 122
E G C+ TC + A C C G+ G DC E + C+
Sbjct 389 ---EHDLDDCAGRACANGGTCVEGGGAHRCSCALGFGGRDCRERADPCA 434
Score = 27.7 bits (60), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 11/17 (64%), Positives = 12/17 (70%), Gaps = 0/17 (0%)
Query 64 GKCTCLAGWGGPQCIVP 80
G+C CL GW GP C VP
Sbjct 235 GECRCLEGWTGPLCTVP 251
> Hs21450863
Length=1272
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 18/32 (56%), Positives = 21/32 (65%), Gaps = 3/32 (9%)
Query 53 CSNDGKCDPATGKCTCLAGWGGPQCIVPQPCG 84
C N G+C+P TG+C C AGW G QC Q CG
Sbjct 105 CVNGGRCNPGTGQCVCPAGWVGEQC---QHCG 133
> Hs21450861
Length=1429
Score = 37.0 bits (84), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 18/32 (56%), Positives = 21/32 (65%), Gaps = 3/32 (9%)
Query 53 CSNDGKCDPATGKCTCLAGWGGPQCIVPQPCG 84
C N G+C+P TG+C C AGW G QC Q CG
Sbjct 105 CVNGGRCNPGTGQCVCPAGWVGEQC---QHCG 133
> CE14412
Length=1372
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 33/102 (32%), Positives = 42/102 (41%), Gaps = 16/102 (15%)
Query 33 CECTPEFPAANC---VDLCSGVECSNDGKCDPATG--KCTCLAGWGGPQCI------VPQ 81
C C P + C D C+ C ND +C G KC C +GW GP+C +
Sbjct 204 CVCKPGYTGPRCDVKQDQCADSPCLNDAQCVDMGGAYKCVCKSGWTGPKCEQDNGSCAAK 263
Query 82 PCGTEGAICSGNATCDQEALSCVCKPGYTGEDCTELSPECSG 123
PC G S A CVC PG +G++C C G
Sbjct 264 PCRNNGFCVSLVAD-----YFCVCPPGVSGKNCESAPNRCIG 300
Score = 33.9 bits (76), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 41/137 (29%), Positives = 57/137 (41%), Gaps = 24/137 (17%)
Query 2 SGSNCETHEANC----CESDSDC-NAPNGVCNSSNTCECTPEFPAANC---VDLCSGVEC 53
+G NC+ + C C++D+ C + NG CEC + +C VD C+ C
Sbjct 95 TGENCDQNIDECAASPCQNDAKCIDEINGY-----MCECADGYEGVHCQHLVDHCAKQPC 149
Query 54 SNDGKCDP--ATGKCTCLAGWGGPQCIV------PQPCGTEGAICSGNATCDQEALSCVC 105
N+ C AT C C G+ G C + C G +A D CVC
Sbjct 150 HNNATCTNMGATYHCDCTLGFDGVHCEMNIDECAENQCDKLGTESCRDAVND---FKCVC 206
Query 106 KPGYTGEDCTELSPECS 122
KPGYTG C +C+
Sbjct 207 KPGYTGPRCDVKQDQCA 223
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 36/82 (43%), Gaps = 13/82 (15%)
Query 42 ANCVDLCSGVECSNDGKCDPATGKCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQEAL 101
A CVDL ND KC+ C G+ G +C + + G +
Sbjct 40 ATCVDLI------NDYKCE-------CPTGFSGKRCHIKENLCASSPCVHGLCIDKLYSR 86
Query 102 SCVCKPGYTGEDCTELSPECSG 123
C+C+PG+TGE+C + EC+
Sbjct 87 QCLCQPGWTGENCDQNIDECAA 108
> CE07053
Length=1722
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 48/170 (28%), Positives = 67/170 (39%), Gaps = 24/170 (14%)
Query 1 DSGSNCETHEANC----CESDSDCNAPNGVCNSSNTCECTPEFPAANCV---DLCSGVEC 53
D C+++E +C C+++ C A NG + C C P F +C D C C
Sbjct 117 DGSYICQSNEPSCATHTCQNNGTCVAENG----NVKCACPPGFVGDHCETDEDECKENFC 172
Query 54 SNDGKCDPATG--KCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQEA--LSCVCKPGY 109
N C+ G +C CL G+ G C + C N C LSC C PG+
Sbjct 173 QNGADCENLKGSYECKCLKGFSGKYCEIQDKKQCTSDYCHNNGQCISTGSDLSCKCSPGF 232
Query 110 TGEDCTELSPEC--------SGLCTVGGSVIRDSVSECKVDCYSACCKHL 151
G C EL E + +C++ R + EC A CK L
Sbjct 233 DGAFC-ELKAEVNECICENPAHVCSLVNGTSRTTQCECPSGFMGADCKEL 281
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 37/127 (29%), Positives = 54/127 (42%), Gaps = 15/127 (11%)
Query 3 GSNCETHEANCCESDSDCNAPNGVCNSSN--TCECTPEFPAANC---VD-LCSGVECSND 56
G++C+ +A C+ + N + V + N TC C P F C VD L +G +C N
Sbjct 275 GADCKELQARPCDREPCLNGGHCVDDGQNLFTCFCLPSFTGIYCGEPVDCLVNGSDCKNG 334
Query 57 GKCDPATG--KCTCLAGWGGPQCIVPQPCGTE----GAICSGNATCDQEAL---SCVCKP 107
GKC A C C G+ G C + + C +C +A CVC+
Sbjct 335 GKCVFALAATTCQCPEGFNGSNCEISNSYRSHPTCSDIRCLNGGSCKLDAEGEPFCVCEE 394
Query 108 GYTGEDC 114
G+ G C
Sbjct 395 GFDGPFC 401
Score = 28.9 bits (63), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 28/107 (26%), Positives = 41/107 (38%), Gaps = 26/107 (24%)
Query 52 ECSN-----DGKCDPATG--KCTCLAGWGGPQCIVPQPCGTEGA---------------- 88
EC+N +G+C G KC C G+ GP+C V PC + A
Sbjct 1561 ECANPNNCINGECTNTLGNYKCACRNGFIGPRCSVRNPCTAQIASNNISSVTCVHGKCVN 1620
Query 89 -ICSGNATCDQEALSCVCKPGYTGEDCTELSPE--CSGLCTVGGSVI 132
+ + C C GYTG C++ E S + + G +I
Sbjct 1621 PVVQIEKNREVAKYECACDRGYTGPTCSQRIKESAMSNISYLFGPII 1667
> CE10670
Length=1827
Score = 36.2 bits (82), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 37/123 (30%), Positives = 55/123 (44%), Gaps = 18/123 (14%)
Query 11 ANCCESDSDCNAPNGVCNSSNTCECTPEFPAANCV---DLCSGVECSNDGKCDPA--TGK 65
A CE++++C+ V C C P + C D+CS C N GKC+ T K
Sbjct 1675 AKMCENNAECS----VFMHRAQCHCKPGYVGDRCEMLEDVCSTQPCYNGGKCEQVGTTYK 1730
Query 66 CTCLAGWGGPQCIVPQPCGTE--GAICSGNATCDQ----EALSCVCKPGYTGEDCTELSP 119
CTC + G +C Q E G C C ++ +C+C+ G+ G C E++
Sbjct 1731 CTCPKMFNGARC---QFESDECNGVKCPNGGVCHDLPGVKSTTCLCRTGFAGPQCEEITD 1787
Query 120 ECS 122
CS
Sbjct 1788 ICS 1790
Score = 35.4 bits (80), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 34/97 (35%), Positives = 42/97 (43%), Gaps = 12/97 (12%)
Query 29 SSNTCECTPEFPAANCV---DLCSGVECSNDGKCDPATG----KCTCLAGWGGPQCI-VP 80
++ C C F A C D C+GV+C N G C G C C G+ GPQC +
Sbjct 1727 TTYKCTCPKMFNGARCQFESDECNGVKCPNGGVCHDLPGVKSTTCLCRTGFAGPQCEEIT 1786
Query 81 QPCGTEGAICSGNATCDQEAL---SCVCKPGYTGEDC 114
C T C A C E L C C PG+ G +C
Sbjct 1787 DICSTNNP-CRNGARCIGEKLGRFKCQCVPGWEGPNC 1822
> Hs21361989
Length=1316
Score = 36.2 bits (82), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 42/123 (34%), Positives = 56/123 (45%), Gaps = 16/123 (13%)
Query 3 GSNCETHEANCCESDSDCNAPNGVCNSS---NTCECTPEFPAANC---VDLCSGVECSND 56
G +CET E N C+S+ N N VC C+C P F C VD C C N
Sbjct 1068 GLHCET-EVNECQSNPCLN--NAVCEDQVGGFLCKCPPGFLGTRCGKNVDECLSQPCKNG 1124
Query 57 GKCDPA--TGKCTCLAGWGGPQC-IVPQPCGTEGAICSGNATCDQE--ALSCVCKPGYTG 111
C + +C C AG+ G C + C + C ATC E + SC C+PG++G
Sbjct 1125 ATCKDGANSFRCLCAAGFTGSHCELNINEC--QSNPCRNQATCVDELNSYSCKCQPGFSG 1182
Query 112 EDC 114
+ C
Sbjct 1183 KRC 1185
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 39/123 (31%), Positives = 53/123 (43%), Gaps = 16/123 (13%)
Query 14 CESDSDCNAP-----NGVCNS---SNTCECTPEFPAANC---VDLCSGVECSNDGKC-DP 61
CE+D D +P NGVC CEC + C ++ CS C N G C D
Sbjct 995 CETDIDECSPLPCLNNGVCKDLVGEFICECPSGYTGQRCEENINECSSSPCLNKGICVDG 1054
Query 62 ATG-KCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQEA--LSCVCKPGYTGEDCTELS 118
G +CTC+ G+ G C + + C NA C+ + C C PG+ G C +
Sbjct 1055 VAGYRCTCVKGFVGLHCET-EVNECQSNPCLNNAVCEDQVGGFLCKCPPGFLGTRCGKNV 1113
Query 119 PEC 121
EC
Sbjct 1114 DEC 1116
> Hs20538457
Length=2570
Score = 36.2 bits (82), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 25/58 (43%), Positives = 28/58 (48%), Gaps = 3/58 (5%)
Query 64 GKCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQEALSCVCKPGYT--GEDCTELSP 119
G C C AG+ GP C P E C N C EA SC C PGYT G +C +P
Sbjct 179 GSCLCFAGYTGPHCDQELPVCQE-LRCPQNTQCSAEAPSCRCLPGYTQQGSECRAPNP 235
Score = 28.5 bits (62), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 25/76 (32%), Positives = 31/76 (40%), Gaps = 19/76 (25%)
Query 48 CSGVECSNDGKCDPA---TGKCTCLAGWGGPQCIVPQ--------PCGTEGAICSGNATC 96
C C+ G+CD +G C C GW GP+C V PC E +GN
Sbjct 2018 CQACRCTVHGRCDEGLGGSGSCFCDEGWTGPRCEVQLELQPVCTPPCAPEAVCRAGN--- 2074
Query 97 DQEALSCVCKPGYTGE 112
SC C GY G+
Sbjct 2075 -----SCECSLGYEGD 2085
> Hs12225240
Length=2570
Score = 35.8 bits (81), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 25/58 (43%), Positives = 28/58 (48%), Gaps = 3/58 (5%)
Query 64 GKCTCLAGWGGPQCIVPQPCGTEGAICSGNATCDQEALSCVCKPGYT--GEDCTELSP 119
G C C AG+ GP C P E C N C EA SC C PGYT G +C +P
Sbjct 179 GSCLCFAGYTGPHCDQELPVCQE-LRCPQNTQCSAEAPSCRCLPGYTQQGSECRAPNP 235
Lambda K H
0.316 0.133 0.466
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1997677330
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40