bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2996_orf2
Length=196
Score E
Sequences producing significant alignments: (Bits) Value
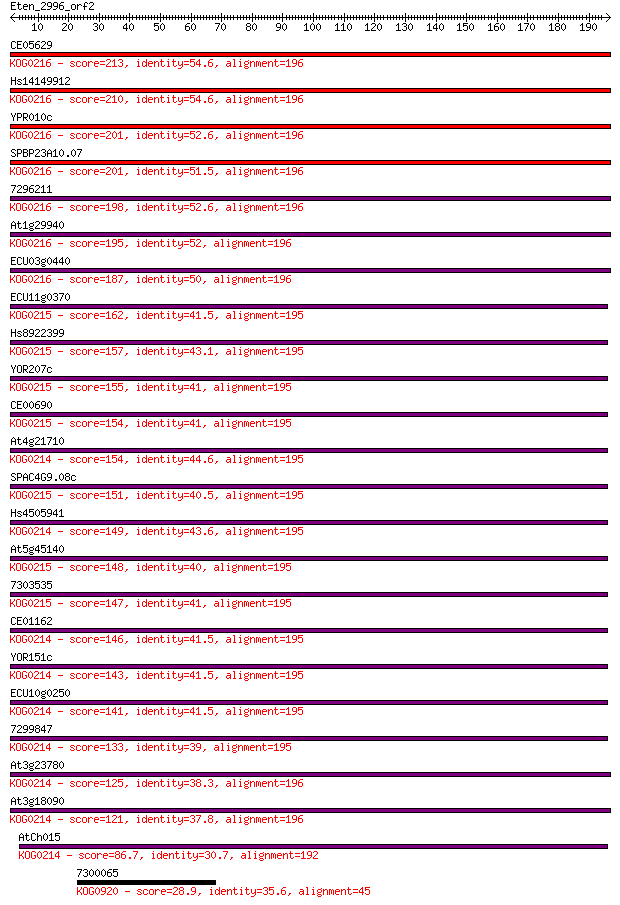
CE05629 213 2e-55
Hs14149912 210 1e-54
YPR010c 201 6e-52
SPBP23A10.07 201 1e-51
7296211 198 6e-51
At1g29940 195 5e-50
ECU03g0440 187 1e-47
ECU11g0370 162 4e-40
Hs8922399 157 9e-39
YOR207c 155 6e-38
CE00690 154 9e-38
At4g21710 154 1e-37
SPAC4G9.08c 151 1e-36
Hs4505941 149 5e-36
At5g45140 148 8e-36
7303535 147 1e-35
CE01162 146 3e-35
YOR151c 143 2e-34
ECU10g0250 141 8e-34
7299847 133 2e-31
At3g23780 125 8e-29
At3g18090 121 1e-27
AtCh015 86.7 3e-17
7300065 28.9 6.2
> CE05629
Length=1127
Score = 213 bits (542), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 107/196 (54%), Positives = 125/196 (63%), Gaps = 37/196 (18%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
R R PIIGDKFASRHGQKGI S LWP E +PFSE+G+VPDI+FNPHGFPSRMTIGM+IE
Sbjct 846 RIERNPIIGDKFASRHGQKGINSFLWPVESLPFSETGMVPDIIFNPHGFPSRMTIGMMIE 905
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
SMAGKAAA HG DA+ F +
Sbjct 906 SMAGKAAATHGENYDASPF-------------------------------------VFNE 928
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
+T +++FG+ L +GY YYG E YSGV G + IF G++YYQRLRHM+ DK QVRA
Sbjct 929 DNTAINHFGELLTKAGYNYYGNETFYSGVDGRQMEMQIFFGIVYYQRLRHMIADKFQVRA 988
Query 181 TGPIDSLTHQPVKGRK 196
TGPID +THQPVKGRK
Sbjct 989 TGPIDPITHQPVKGRK 1004
> Hs14149912
Length=459
Score = 210 bits (534), Expect = 1e-54, Method: Compositional matrix adjust.
Identities = 107/196 (54%), Positives = 129/196 (65%), Gaps = 37/196 (18%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
R R P IGDKFASRHGQKGILS LWP EDMPF+ESG+VPDILFNPHGFPSRMTIGMLIE
Sbjct 231 RVPRNPTIGDKFASRHGQKGILSRLWPAEDMPFTESGMVPDILFNPHGFPSRMTIGMLIE 290
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
SMAGK+AA+HG DAT F + ++E
Sbjct 291 SMAGKSAALHGLCHDATPF-----------------------------------IFSEE- 314
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
++ ++YFG+ L A+GY +YG E LYSG+ G+ L IF G++YYQRLRHMV+DK QVR
Sbjct 315 -NSALEYFGEMLKAAGYNFYGTERLYSGISGLELEADIFIGVVYYQRLRHMVSDKFQVRT 373
Query 181 TGPIDSLTHQPVKGRK 196
TG D +T+QP+ GR
Sbjct 374 TGARDRVTNQPIGGRN 389
> YPR010c
Length=1203
Score = 201 bits (512), Expect = 6e-52, Method: Compositional matrix adjust.
Identities = 103/196 (52%), Positives = 121/196 (61%), Gaps = 37/196 (18%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
R R P IGDKF+SRHGQKG+ S WP DMPFSE+GI PDI+ NPH FPSRMTIGM +E
Sbjct 906 RIRRTPQIGDKFSSRHGQKGVCSRKWPTIDMPFSETGIQPDIIINPHAFPSRMTIGMFVE 965
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
S+AGKA A+HG QD+T WI E
Sbjct 966 SLAGKAGALHGIAQDSTP-------------WIFNE------------------------ 988
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
DTP DYFG+ L +GY Y+G E +YSG G L I+ G++YYQRLRHMV DK QVR+
Sbjct 989 DDTPADYFGEQLAKAGYNYHGNEPMYSGATGEELRADIYVGVVYYQRLRHMVNDKFQVRS 1048
Query 181 TGPIDSLTHQPVKGRK 196
TGP++SLT QPVKGRK
Sbjct 1049 TGPVNSLTMQPVKGRK 1064
> SPBP23A10.07
Length=1227
Score = 201 bits (510), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 101/196 (51%), Positives = 123/196 (62%), Gaps = 37/196 (18%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
R R PIIGDKF+SRHGQKGI S WP DMPF+ESG+ PDI+ NPH FPSRMTIGM IE
Sbjct 944 RITRSPIIGDKFSSRHGQKGICSQKWPTVDMPFTESGMQPDIIINPHAFPSRMTIGMFIE 1003
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
S+AGKA A HG QD+T F + +++Q
Sbjct 1004 SLAGKAGACHGLAQDSTPF-----------------------------------IYSEQQ 1028
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
T DYFG+ L+ +GY Y+G E +YSG+ G + I+ G++YYQRLRHMV+DK QVR
Sbjct 1029 --TAADYFGEQLVKAGYNYHGNEPMYSGITGQEMKADIYIGVVYYQRLRHMVSDKFQVRT 1086
Query 181 TGPIDSLTHQPVKGRK 196
TGPI +LT QPVKGRK
Sbjct 1087 TGPIHNLTRQPVKGRK 1102
> 7296211
Length=1129
Score = 198 bits (504), Expect = 6e-51, Method: Compositional matrix adjust.
Identities = 103/196 (52%), Positives = 125/196 (63%), Gaps = 37/196 (18%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
R R IGDKFASR GQKGI S +P ED+PF+ESG++PDI+FNPHGFPSRMTI M+IE
Sbjct 863 RVPRPATIGDKFASRAGQKGICSQKYPAEDLPFTESGLIPDIVFNPHGFPSRMTIAMMIE 922
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
+MAGK AA+HG DAT FR F ++
Sbjct 923 TMAGKGAAIHGNVYDATPFR-FSEE----------------------------------- 946
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
+T +DYFGK L A GY YYG E LYSGV G +T IF G+++YQRLRHMV DK QVR+
Sbjct 947 -NTAIDYFGKMLEAGGYNYYGTERLYSGVDGREMTADIFFGVVHYQRLRHMVFDKWQVRS 1005
Query 181 TGPIDSLTHQPVKGRK 196
TG +++ THQP+KGRK
Sbjct 1006 TGAVEARTHQPIKGRK 1021
> At1g29940
Length=1114
Score = 195 bits (496), Expect = 5e-50, Method: Compositional matrix adjust.
Identities = 102/197 (51%), Positives = 123/197 (62%), Gaps = 28/197 (14%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFS-ESGIVPDILFNPHGFPSRMTIGMLI 59
R R PIIGDKF+SRHGQKG+ S LWP DMPF+ +G+ PD++ NPH FPSRMTI ML+
Sbjct 824 RHARNPIIGDKFSSRHGQKGVCSQLWPDIDMPFNGVTGMRPDLIINPHAFPSRMTIAMLL 883
Query 60 ESMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQE 119
ES+A K ++HG F DAT FR+ K KT EEE K+
Sbjct 884 ESIAAKGGSLHGKFVDATPFRDAVK--KTNG---------------------EEESKSS- 919
Query 120 QGDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVR 179
VD G L G+ +YG E LYSG G+ L C IF G +YYQRLRHMV+DK QVR
Sbjct 920 ---LLVDDLGSMLKEKGFNHYGTETLYSGYLGVELKCEIFMGPVYYQRLRHMVSDKFQVR 976
Query 180 ATGPIDSLTHQPVKGRK 196
+TG +D LTHQP+KGRK
Sbjct 977 STGQVDQLTHQPIKGRK 993
> ECU03g0440
Length=1062
Score = 187 bits (476), Expect = 1e-47, Method: Composition-based stats.
Identities = 98/196 (50%), Positives = 119/196 (60%), Gaps = 27/196 (13%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
R VR P+IGDKF+SRHGQKG+ SM WP DMPF+ESG VPDI+ NPH FPSRMTIGMLIE
Sbjct 806 RIVRSPMIGDKFSSRHGQKGVCSMHWPGIDMPFTESGCVPDIIINPHAFPSRMTIGMLIE 865
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
S+AGK + G QD T F++ E+ +EG +E+V+ +E
Sbjct 866 SIAGKVGCLSGNEQDGTVFKKSFLLEQ-------EEGD-------------DEKVRRKE- 904
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
Y L G+ YYG E +YSGV G IF G++YYQRL+HMV DK QVR
Sbjct 905 ------YLCSELRRHGFNYYGNEPMYSGVAGNEFRADIFVGVVYYQRLKHMVGDKFQVRT 958
Query 181 TGPIDSLTHQPVKGRK 196
G + S T QPV GRK
Sbjct 959 KGAVVSTTRQPVGGRK 974
> ECU11g0370
Length=1110
Score = 162 bits (410), Expect = 4e-40, Method: Composition-based stats.
Identities = 81/195 (41%), Positives = 107/195 (54%), Gaps = 41/195 (21%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
R R P IGDKF+SRHGQKG++ +L EDMPF++ GIVPDI+ NPHGFPSRMT+G ++E
Sbjct 868 RQTRVPEIGDKFSSRHGQKGVVGLLVRQEDMPFNDQGIVPDIIMNPHGFPSRMTVGKIVE 927
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
++GKA + G D+TAF+E
Sbjct 928 LISGKAGVLEGQILDSTAFKE--------------------------------------- 948
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
V+ + L+ G+ Y GK+ SG G PL +IF G ++YQRL+HMV DK +RA
Sbjct 949 --NSVEQTCELLIKHGFSYSGKDCFTSGTTGAPLAAYIFFGPVFYQRLKHMVADKIHMRA 1006
Query 181 TGPIDSLTHQPVKGR 195
GP LT QP +GR
Sbjct 1007 RGPRAILTRQPTEGR 1021
> Hs8922399
Length=343
Score = 157 bits (398), Expect = 9e-39, Method: Compositional matrix adjust.
Identities = 84/195 (43%), Positives = 106/195 (54%), Gaps = 41/195 (21%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
R R+P IGDKF+SRHGQKG+ ++ P EDMPF +SGI PDI+ NPHGFPSRMT+G LIE
Sbjct 96 RQTRRPEIGDKFSSRHGQKGVCGLIVPQEDMPFCDSGICPDIIMNPHGFPSRMTVGKLIE 155
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
+AGKA + G F TAF
Sbjct 156 LLAGKAGVLDGRFHYGTAF----------------------------------------- 174
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
G + V + L+ GY Y GK+ + SG+ G PL +I+ G +YYQ+L+HMV DK RA
Sbjct 175 GGSKVKDVCEDLVRHGYNYLGKDYVTSGITGEPLEAYIYFGPVYYQKLKHMVLDKMHARA 234
Query 181 TGPIDSLTHQPVKGR 195
GP LT QP +GR
Sbjct 235 RGPRAVLTRQPTEGR 249
> YOR207c
Length=1149
Score = 155 bits (391), Expect = 6e-38, Method: Compositional matrix adjust.
Identities = 80/195 (41%), Positives = 107/195 (54%), Gaps = 41/195 (21%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
R R+P +GDKF+SRHGQKG+ ++ EDMPF++ GIVPDI+ NPHGFPSRMT+G +IE
Sbjct 901 RQNRRPELGDKFSSRHGQKGVCGIIVKQEDMPFNDQGIVPDIIMNPHGFPSRMTVGKMIE 960
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
++GKA ++G + T F
Sbjct 961 LISGKAGVLNGTLEYGTCF----------------------------------------- 979
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
G + ++ K L+ G+ Y GK+ LYSG+ G L +IF G IYYQ+L+HMV DK RA
Sbjct 980 GGSKLEDMSKILVDQGFNYSGKDMLYSGITGECLQAYIFFGPIYYQKLKHMVLDKMHARA 1039
Query 181 TGPIDSLTHQPVKGR 195
GP LT QP +GR
Sbjct 1040 RGPRAVLTRQPTEGR 1054
> CE00690
Length=1207
Score = 154 bits (390), Expect = 9e-38, Method: Compositional matrix adjust.
Identities = 80/195 (41%), Positives = 108/195 (55%), Gaps = 41/195 (21%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
R R+P +GDKF+SRHGQKG+ ++ EDMPF++ G+VPD++ NPHG+PSRMT+G L+E
Sbjct 960 RQTRRPELGDKFSSRHGQKGVCGLIAQQEDMPFNDLGMVPDMIMNPHGYPSRMTVGKLME 1019
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
++GKA V+G + TAF
Sbjct 1020 LLSGKAGVVNGTYHYGTAF----------------------------------------- 1038
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
G V + L A GY Y GK+ L SG+ G PL+ +I+ G IYYQ+L+HMV DK RA
Sbjct 1039 GGDQVKDVCEELAACGYNYMGKDMLTSGITGQPLSAYIYFGPIYYQKLKHMVLDKMHARA 1098
Query 181 TGPIDSLTHQPVKGR 195
GP +LT QP +GR
Sbjct 1099 RGPRAALTRQPTEGR 1113
> At4g21710
Length=1188
Score = 154 bits (389), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 87/195 (44%), Positives = 100/195 (51%), Gaps = 41/195 (21%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
RSVR P IGDKF+SRHGQKG + M + EDMP++ G+ PDI+ NPH PSRMTIG LIE
Sbjct 930 RSVRIPQIGDKFSSRHGQKGTVGMTYTQEDMPWTIEGVTPDIIVNPHAIPSRMTIGQLIE 989
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
+ GK AA G DAT F
Sbjct 990 CIMGKVAAHMGKEGDATPF----------------------------------------- 1008
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
D VD KAL GYQ G E +Y+G G PLT IF G YYQRL+HMV DK R
Sbjct 1009 TDVTVDNISKALHKCGYQMRGFERMYNGHTGRPLTAMIFLGPTYYQRLKHMVDDKIHSRG 1068
Query 181 TGPIDSLTHQPVKGR 195
GP+ LT QP +GR
Sbjct 1069 RGPVQILTRQPAEGR 1083
> SPAC4G9.08c
Length=1165
Score = 151 bits (381), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 79/195 (40%), Positives = 103/195 (52%), Gaps = 41/195 (21%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
R R+P +GDKF+SRHGQKG+ ++ EDMPF++ GI PDI+ NPHGFPSRMT+G +IE
Sbjct 917 RQTRRPELGDKFSSRHGQKGVCGVIVQQEDMPFNDQGICPDIIMNPHGFPSRMTVGKMIE 976
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
++GK + G + T F
Sbjct 977 LLSGKVGVLRGTLEYGTCF----------------------------------------- 995
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
G T V+ + L+ GY Y GK+ L SG+ G L +IF G IYYQ+L+HMV DK RA
Sbjct 996 GGTKVEDASRILVEHGYNYSGKDMLTSGITGETLEAYIFMGPIYYQKLKHMVMDKMHARA 1055
Query 181 TGPIDSLTHQPVKGR 195
GP LT QP +GR
Sbjct 1056 RGPRAVLTRQPTEGR 1070
> Hs4505941
Length=1174
Score = 149 bits (375), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 85/195 (43%), Positives = 99/195 (50%), Gaps = 40/195 (20%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
RSVR P IGDKFASRHGQKG + + EDMPF+ GI PDI+ NPH PSRMTIG LIE
Sbjct 924 RSVRIPQIGDKFASRHGQKGTCGIQYRQEDMPFTCEGITPDIIINPHAIPSRMTIGHLIE 983
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
+ GK +A G DAT F + V Q+
Sbjct 984 CLQGKVSANKGEIGDATPF--------------------------------NDAVNVQKI 1011
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
+ DY GY G E LY+G G +T IF G YYQRL+HMV DK RA
Sbjct 1012 SNLLSDY--------GYHLRGNEVLYNGFTGRKITSQIFIGPTYYQRLKHMVDDKIHSRA 1063
Query 181 TGPIDSLTHQPVKGR 195
GPI L QP++GR
Sbjct 1064 RGPIQILNRQPMEGR 1078
> At5g45140
Length=1194
Score = 148 bits (373), Expect = 8e-36, Method: Compositional matrix adjust.
Identities = 78/195 (40%), Positives = 104/195 (53%), Gaps = 36/195 (18%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
R R+P +GDKF+SRHGQKG+ ++ ED PFSE GI PD++ NPHGFPSRMT+G +IE
Sbjct 915 RHTRRPELGDKFSSRHGQKGVCGIIIQQEDFPFSELGICPDLIMNPHGFPSRMTVGKMIE 974
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
+ KA G F +AF E + GH
Sbjct 975 LLGSKAGVSCGRFHYGSAFGE-------------RSGHAD-------------------- 1001
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
V+ L+ G+ Y GK+ LYSG+ G P+ +IF G IYYQ+L+HMV DK R
Sbjct 1002 ---KVETISATLVEKGFSYSGKDLLYSGISGEPVEAYIFMGPIYYQKLKHMVLDKMHARG 1058
Query 181 TGPIDSLTHQPVKGR 195
+GP +T QP +G+
Sbjct 1059 SGPRVMMTRQPTEGK 1073
> 7303535
Length=1137
Score = 147 bits (372), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 80/195 (41%), Positives = 107/195 (54%), Gaps = 41/195 (21%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
R R P IGDKF+SRHGQKG+ ++ EDMPF++ GI PD++ NPHGFPSRMT+G +E
Sbjct 892 RQTRIPEIGDKFSSRHGQKGVTGLIVEQEDMPFNDFGICPDMIMNPHGFPSRMTVGKTLE 951
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
+ GKA + G F TAF GG + E+++A+
Sbjct 952 LLGGKAGLLEGKFHYGTAF-------------------------GG---SKVEDIQAE-- 981
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
L G+ Y GK+ YSG+ G PL +I++G +YYQ+L+HMV DK RA
Sbjct 982 -----------LERHGFNYVGKDFFYSGITGTPLEAYIYSGPVYYQKLKHMVQDKMHARA 1030
Query 181 TGPIDSLTHQPVKGR 195
GP LT QP +GR
Sbjct 1031 RGPKAVLTRQPTQGR 1045
> CE01162
Length=1193
Score = 146 bits (368), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 81/195 (41%), Positives = 98/195 (50%), Gaps = 40/195 (20%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
RSVR P IGDKFASRHGQKG + +++ EDMPF+ G+ PDI+ NPH PSRMTIG LIE
Sbjct 930 RSVRLPQIGDKFASRHGQKGTMGIMYRQEDMPFTAEGLTPDIIINPHAVPSRMTIGHLIE 989
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
+ GK +A G DAT F + +K
Sbjct 990 CLQGKLSANKGEIGDATPFNDTVNVQKISG------------------------------ 1019
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
L GY G E +Y+G G LT IF G YYQRL+HMV DK RA
Sbjct 1020 ----------LLCEYGYHLRGNEVMYNGHTGKKLTTQIFFGPTYYQRLKHMVDDKIHSRA 1069
Query 181 TGPIDSLTHQPVKGR 195
GPI + QP++GR
Sbjct 1070 RGPIQMMNRQPMEGR 1084
> YOR151c
Length=1224
Score = 143 bits (361), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 81/195 (41%), Positives = 98/195 (50%), Gaps = 41/195 (21%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
R+ + P IGDKFASRHGQKG + + + EDMPF+ GIVPD++ NPH PSRMT+ LIE
Sbjct 969 RTTKIPQIGDKFASRHGQKGTIGITYRREDMPFTAEGIVPDLIINPHAIPSRMTVAHLIE 1028
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
+ K AA+ G DA+ F
Sbjct 1029 CLLSKVAALSGNEGDASPF----------------------------------------- 1047
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
D V+ K L GYQ G E +Y+G G L IF G YYQRLRHMV DK RA
Sbjct 1048 TDITVEGISKLLREHGYQSRGFEVMYNGHTGKKLMAQIFFGPTYYQRLRHMVDDKIHARA 1107
Query 181 TGPIDSLTHQPVKGR 195
GP+ LT QPV+GR
Sbjct 1108 RGPMQVLTRQPVEGR 1122
> ECU10g0250
Length=1141
Score = 141 bits (356), Expect = 8e-34, Method: Compositional matrix adjust.
Identities = 81/195 (41%), Positives = 98/195 (50%), Gaps = 41/195 (21%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
RS R P +GDKFASRH QKG + + EDMPF+ GIVPDI+ NPH PSRMTIG LIE
Sbjct 893 RSGRIPQMGDKFASRHAQKGTIGITLRQEDMPFTSEGIVPDIIINPHAIPSRMTIGHLIE 952
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
+ GK +A+ G DAT F
Sbjct 953 CLLGKVSAMSGEEGDATPF----------------------------------------- 971
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
VD L + G+Q G E +Y+G+ G L +F G YYQRL+HMV DK RA
Sbjct 972 SGVTVDGISSRLKSYGFQQRGLEVMYNGMTGRKLRAQMFFGPTYYQRLKHMVDDKIHARA 1031
Query 181 TGPIDSLTHQPVKGR 195
GP+ LT QPV+GR
Sbjct 1032 RGPLQILTRQPVEGR 1046
> 7299847
Length=1176
Score = 133 bits (335), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 76/195 (38%), Positives = 93/195 (47%), Gaps = 40/195 (20%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
RSVR P IGDKFASRHGQKG + + EDM F+ G+ PDI+ NPH PSRMTIG LIE
Sbjct 926 RSVRIPQIGDKFASRHGQKGTCGIQYRQEDMAFTCEGLAPDIIINPHAIPSRMTIGHLIE 985
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
+ GK + G DAT F + +K
Sbjct 986 CLQGKLGSNKGEIGDATPFNDAVNVQKIST------------------------------ 1015
Query 121 GDTPVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRA 180
L GY G E +Y+G G + +F G YYQRL+HMV DK RA
Sbjct 1016 ----------FLQEYGYHLRGNEVMYNGHTGRKINAQVFLGPTYYQRLKHMVDDKIHSRA 1065
Query 181 TGPIDSLTHQPVKGR 195
GP+ L QP++GR
Sbjct 1066 RGPVQILVRQPMEGR 1080
> At3g23780
Length=946
Score = 125 bits (313), Expect = 8e-29, Method: Compositional matrix adjust.
Identities = 75/197 (38%), Positives = 97/197 (49%), Gaps = 32/197 (16%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
R VR P +GDKF+S HGQKG+L L ++ PF+ GIVPDI+ NPH FPSR T G L+E
Sbjct 677 RQVRSPCLGDKFSSMHGQKGVLGYLEEQQNFPFTIQGIVPDIVINPHAFPSRQTPGQLLE 736
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
+ K A I +EG + R+ TP
Sbjct 737 AALSKGIACP----------------------IQKEGSSAAYTKLTRHATP--------- 765
Query 121 GDTP-VDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVR 179
TP V + L +G+ +G E +Y+G G + IF G +YQRL HM DK + R
Sbjct 766 FSTPGVTEITEQLHRAGFSRWGNERVYNGRSGEMMRSMIFMGPTFYQRLVHMSEDKVKFR 825
Query 180 ATGPIDSLTHQPVKGRK 196
TGP+ LT QPV RK
Sbjct 826 NTGPVHPLTRQPVADRK 842
> At3g18090
Length=1038
Score = 121 bits (303), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 74/197 (37%), Positives = 100/197 (50%), Gaps = 31/197 (15%)
Query 1 RSVRKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIE 60
R VR P +GDKF+S HGQKG+L L ++ PF+ GIVPDI+ NPH FPSR T G L+E
Sbjct 768 RQVRSPCLGDKFSSMHGQKGVLGYLEEQQNFPFTIQGIVPDIVINPHAFPSRQTPGQLLE 827
Query 61 SMAGKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQ 120
+ K A +K+E + A + + R+ TP
Sbjct 828 AALSKGIAC-----------PIQKKEGSSAAYT----------KLTRHATP--------- 857
Query 121 GDTP-VDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVR 179
TP V + L +G+ +G E +Y+G G + IF G +YQRL HM +K + R
Sbjct 858 FSTPGVTEITEQLHRAGFSRWGNERVYNGRSGEMMRSLIFMGPTFYQRLVHMSENKVKFR 917
Query 180 ATGPIDSLTHQPVKGRK 196
TGP+ LT QPV RK
Sbjct 918 NTGPVHPLTRQPVADRK 934
> AtCh015
Length=1072
Score = 86.7 bits (213), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 59/192 (30%), Positives = 87/192 (45%), Gaps = 26/192 (13%)
Query 4 RKPIIGDKFASRHGQKGILSMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIESMA 63
R+ +GDK A RHG KGI+S + P +DMP+ + G D++FNP G PSRM +G + E
Sbjct 814 REIKVGDKVAGRHGNKGIISKILPRQDMPYLQDGRPVDMVFNPLGVPSRMNVGQIFECSL 873
Query 64 GKAAAVHGAFQDATAFREFRKQEKTGARWIDQEGHHGYVLRGGRYRTPEEEVKAQEQGDT 123
G A ++ F E +QE + R E +A +Q
Sbjct 874 GLAGSLLDRHYRIAPFDERYEQEAS------------------RKLVFSELYEASKQTAN 915
Query 124 PVDYFGKALLASGYQYYGKEELYSGVYGMPLTCHIFTGLIYYQRLRHMVTDKAQVRATGP 183
P + +Y GK ++ G G P + G Y +L H V DK R++G
Sbjct 916 PWVF--------EPEYPGKSRIFDGRTGDPFEQPVIIGKPYILKLIHQVDDKIHGRSSGH 967
Query 184 IDSLTHQPVKGR 195
+T QP++GR
Sbjct 968 YALVTQQPLRGR 979
> 7300065
Length=1434
Score = 28.9 bits (63), Expect = 6.2, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 28/45 (62%), Gaps = 5/45 (11%)
Query 23 SMLWPHEDMPFSESGIVPDILFNPHGFPSRMTIGMLIESMAGKAA 67
S++W ED+ + VP+I N HG+ + + I ++I++M KAA
Sbjct 313 SIIWNEEDVGHQQ---VPEI--NKHGYRAAVKIIVIIDNMERKAA 352
Lambda K H
0.321 0.138 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3335884518
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40