bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2993_orf2
Length=358
Score E
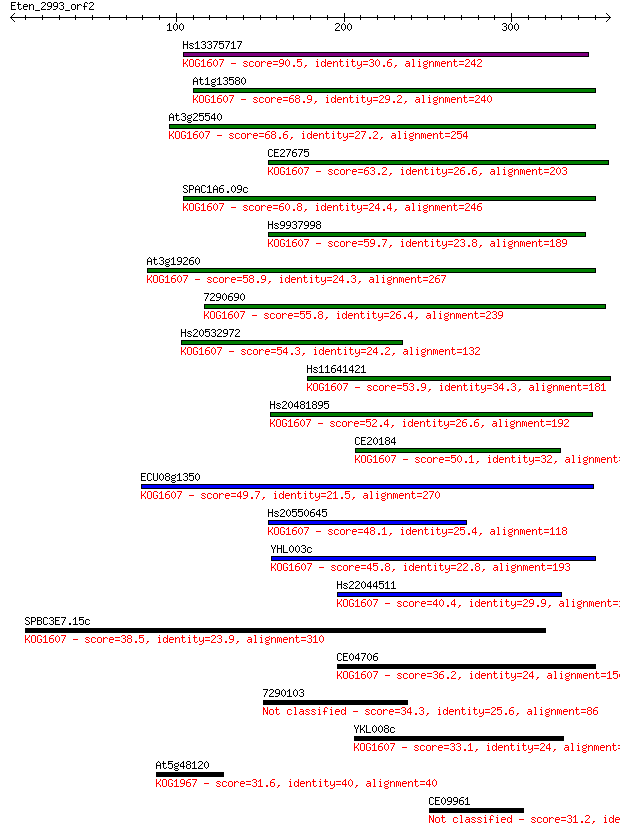
Sequences producing significant alignments: (Bits) Value
Hs13375717 90.5 5e-18
At1g13580 68.9 2e-11
At3g25540 68.6 2e-11
CE27675 63.2 9e-10
SPAC1A6.09c 60.8 4e-09
Hs9937998 59.7 1e-08
At3g19260 58.9 1e-08
7290690 55.8 1e-07
Hs20532972 54.3 4e-07
Hs11641421 53.9 6e-07
Hs20481895 52.4 2e-06
CE20184 50.1 7e-06
ECU08g1350 49.7 1e-05
Hs20550645 48.1 3e-05
YHL003c 45.8 2e-04
Hs22044511 40.4 0.006
SPBC3E7.15c 38.5 0.024
CE04706 36.2 0.12
7290103 34.3 0.43
YKL008c 33.1 1.1
At5g48120 31.6 2.9
CE09961 31.2 4.0
> Hs13375717
Length=394
Score = 90.5 bits (223), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 74/250 (29%), Positives = 121/250 (48%), Gaps = 19/250 (7%)
Query 104 RWSEQVRQLLCSHFGEMTWKMLYFLMTTAVCLLSFRNERW-WPQQLGGEGAEEQLWQGYP 162
R Q R L F E +W+ L++L + L +E W W A W YP
Sbjct 120 RRRNQDRPQLTKKFCEASWRFLFYLSSFVGGLSVLYHESWLW--------APVMCWDRYP 171
Query 163 MQQNSVYCHLYFYVAFGHHLASLIYILKSPWLPDFFDRLLPCAAALCLIYFSYMSNFLRV 222
Q + ++ + G +L+ LI + DF ++++ A+ L+ FSY +N LR+
Sbjct 172 NQTLKPSLYWWYLLELGFYLSLLIRLPFDVKRKDFKEQVIHHFVAVILMTFSYSANLLRI 231
Query 223 GVVILFCHDICDIFTCGCKAFVDTPYHKVTIGLFVLLTVCWFYFRLYTFPSAALFPI-YK 281
G ++L HD D CK Y +V LF++ + +FY RL FP+ L+ Y+
Sbjct 232 GSLVLLLHDSSDYLLEACKMVNYMQYQQVCDALFLIFSFVFFYTRLVLFPTQILYTTYYE 291
Query 282 AIKTRPDHSETEGSSYFIFILLTLSLMNIYWFVLMVKMFVHFILSGQM-RDLHSRV---- 336
+I R G +F +L+ L L++++W L+++M F+ GQM +D+ S V
Sbjct 292 SISNR---GPFFGYYFFNGLLMLLQLLHVFWSCLILRMLYSFMKKGQMEKDIRSDVEESD 348
Query 337 -SEEVSSAQS 345
SEEV++AQ
Sbjct 349 SSEEVAAAQE 358
> At1g13580
Length=308
Score = 68.9 bits (167), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 70/247 (28%), Positives = 108/247 (43%), Gaps = 16/247 (6%)
Query 110 RQLLCSHFGEMTWKMLYFLMTTAVCLLSFRNERWWPQ-QLGGEGAEEQLWQGYPMQQNSV 168
R+ F E WK +Y+L + L NE W+ + G +Q W P QQ +
Sbjct 68 RKKKIRKFKESAWKCVYYLSAEILALSVTYNEPWFMNTKYFWVGPGDQTW---PDQQTKL 124
Query 169 YCHL-YFYVAFGHHLASLIYILKSPWLPDFFDRLLPCAAALCLIYFSYMSNFLRVGVVIL 227
L Y +VA + + + DF + A L LI SY+ +F RVG V+L
Sbjct 125 KLKLLYMFVAGFYTYSIFALVFWETRRSDFGVSMGHHIATLILIVLSYVCSFSRVGSVVL 184
Query 228 FCHDICDIFTCGCKAFVDTPYHKVTIGLFVLLTVCWFYFRLYTFPSAALFPI-YKAI-KT 285
HD D+F K + ++ F+L + W RL +P L+ Y+ + +
Sbjct 185 ALHDASDVFLEVGKMSKYSGAERIASFSFILFVLSWIILRLIYYPFWILWSTSYEVVLEL 244
Query 286 RPDHSETEGSSY---FIFILLTLSLMNIYWFVLMVKMFVHFILSGQMRDLHSRVSEEVSS 342
D EG Y F +L L +++IYW+VLM +M V Q++D ++SE+V S
Sbjct 245 DKDKHPIEGPIYYYMFNTLLYCLLVLHIYWWVLMYRMLVK-----QIQD-RGKLSEDVRS 298
Query 343 AQSRSRE 349
E
Sbjct 299 DSEGEDE 305
> At3g25540
Length=310
Score = 68.6 bits (166), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 69/267 (25%), Positives = 115/267 (43%), Gaps = 34/267 (12%)
Query 96 ADRLLPRDRWSEQVRQLLCSHFGEMTWKMLYFLMTTAVCLLSFRNERWWPQQL-GGEGAE 154
+D + R + S +VR+ F E WK +Y+L + L NE W+ L G
Sbjct 62 SDNIKDRKKNSPKVRK-----FKESAWKCIYYLSAELLALSVTYNEPWFSNTLYFWIGPG 116
Query 155 EQLWQGYPMQQNSVYCHLYFYVAFGHHLASLIYILKSPWLPDFFDRLLPCAAALCLIYFS 214
+Q+W PM+ + +++ + + + +L++ DF + L LI S
Sbjct 117 DQIWPDQPMKMKLKFLYMFAAGFYTYSIFALVFWETRR--SDFGVSMGHHITTLVLIVLS 174
Query 215 YMSNFLRVGVVILFCHDICDIFT--------CGCKAFVDTPYHKVTIGLFVLLTVCWFYF 266
Y+ R G VIL HD D+F CG ++ FVL + W
Sbjct 175 YICRLTRAGSVILALHDASDVFLEIGKMSKYCGAESLASIS--------FVLFALSWVVL 226
Query 267 RLYTFPSAALFPI-YKAIKTRPDHSETEGSS-YFIF--ILLTLSLMNIYWFVLMVKMFVH 322
RL +P L+ Y+ I T G Y++F +L L +++I+W+VL+ +M V
Sbjct 227 RLIYYPFWILWSTSYQIIMTVDKEKHPNGPILYYMFNTLLYFLLVLHIFWWVLIYRMLVK 286
Query 323 FILSGQMRDLHSRVSEEVSSAQSRSRE 349
Q++D ++SE+V S E
Sbjct 287 -----QVQD-RGKLSEDVRSDSESDDE 307
> CE27675
Length=368
Score = 63.2 bits (152), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 54/217 (24%), Positives = 92/217 (42%), Gaps = 14/217 (6%)
Query 155 EQLWQGYPMQQNSVYCHLYFYVAFGHHLASLIYILKSPWLPDFFDRLLPCAAALCLIYFS 214
+Q W GYP Y+ + G + + LI DF+ ++ + L+ S
Sbjct 123 KQCWIGYPFHPVPDTIWWYYMIETGFYYSLLIGSTFDVRRSDFWQLMVHHVITIFLLSSS 182
Query 215 YMSNFLRVGVVILFCHDICDIFTCGCKAF-VDTPYHKVTIGLFVLLTVCWFYFRLYTFPS 273
+ NF+RVG +IL HD+ D+F G K D +T +FVL W RL +P
Sbjct 183 WTINFVRVGTLILLSHDVSDVFLEGGKLVRYDAHNKNMTNFMFVLFFSSWVATRLIYYPF 242
Query 274 AALFPIYK--AIKTRPDH-------SETEGSSYFIFILLTLSLMNIYWFVLMVKMFVHFI 324
+ A +PD+ S +F L+ L ++I+W +++++
Sbjct 243 IVIRSAVTEAAALIQPDYILWDYQLSPPYAPRLIVFALILLFFLHIFWTFIILRIAYRTS 302
Query 325 LSGQMRDLH----SRVSEEVSSAQSRSREISSKASKA 357
GQ +D+ S EE + + R+R + K +K
Sbjct 303 TGGQAKDVRSDSDSDYDEEEMARRERTRLLKKKKNKV 339
> SPAC1A6.09c
Length=390
Score = 60.8 bits (146), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 60/255 (23%), Positives = 95/255 (37%), Gaps = 17/255 (6%)
Query 104 RWSEQVRQLLCSHFGEMTWKMLYFLMTTAVCLLSFRNERWWPQQLGGEGAEEQLWQGYPM 163
W + R+++ F E + Y+L + L +R+ +W EE+L++ YP
Sbjct 143 NWGVRNRKVII-RFCEQGYSFFYYLCFWFLGLYIYRSSNYWSN-------EEKLFEDYPQ 194
Query 164 QQNSVYCHLYFYVAFGHHLASLIYILKSPWLPDFFDRLLPCAAALCLIYFSYMSNFLRVG 223
S Y+ + G L ++ + D + LI SY NFLRVG
Sbjct 195 YYMSPLFKAYYLIQLGFWLQQILVLHLEQRRADHWQMFAHHIVTCALIILSYGFNFLRVG 254
Query 224 VVILFCHDICDIFTCGCKAFVDTPYHKVTIGLFVLLTVCWFYFRLYTFPSAALFPIYKAI 283
IL+ D+ D G K + K+ LF + W Y R Y F + A
Sbjct 255 NAILYIFDLSDYILSGGKMLKYLGFGKICDYLFGIFVASWVYSRHYLFSKILRVVVTNAP 314
Query 284 KTRPDHSETEGSSY---------FIFILLTLSLMNIYWFVLMVKMFVHFILSGQMRDLHS 334
+ + Y FI +L TL L+ WF ++VK+ + D S
Sbjct 315 EIIGGFHLDVPNGYIFNKPIYIAFIILLFTLQLLIYIWFGMIVKVAYRVFSGEEATDSRS 374
Query 335 RVSEEVSSAQSRSRE 349
E A S + +
Sbjct 375 DDEGEDEEASSTNED 389
> Hs9937998
Length=230
Score = 59.7 bits (143), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 45/191 (23%), Positives = 84/191 (43%), Gaps = 8/191 (4%)
Query 155 EQLWQGYPMQQNSVYCHLYFYVAFGHHLASLIYILKSPWLPDFFDRLLPCAAALCLIYFS 214
+++W+GYP+Q + Y+ + + + L I DF ++++ A + LI FS
Sbjct 14 KKVWEGYPIQSTIPSQYWYYMIELSFYWSLLFSIASDVKRKDFKEQIIHHVATIILISFS 73
Query 215 YMSNFLRVGVVILFCHDICDIFTCGCKAFVDTPYHKVTIGLFVLLTVCWFYFRLYTFPSA 274
+ +N++R G +I+ HD D K F + +F++ + + RL P
Sbjct 74 WFANYIRAGTLIMALHDSSDYLLESAKMFNYAGWKNTCNNIFIVFAIVFIITRLVILP-- 131
Query 275 ALFPIYKAIKTRPDHSETEGSSYFIF--ILLTLSLMNIYWFVLMVKMFVHFILSGQMRDL 332
F I P Y+ F ++ L L++I+W L+++M FI + D
Sbjct 132 --FWILHCTLVYPLELYPAFFGYYFFNSMMGVLQLLHIFWAYLILRMAHKFITGKLVED- 188
Query 333 HSRVSEEVSSA 343
R EV+ +
Sbjct 189 -ERSDREVTES 198
> At3g19260
Length=296
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 65/275 (23%), Positives = 115/275 (41%), Gaps = 29/275 (10%)
Query 83 LFQGLCTWILKPLADRLLPRDRWSEQVRQLLCSHFGEMTWKMLYFLMTTAVCLLSFRNER 142
+FQ + W+L + + D + + + + C E WK+LY+ L +E
Sbjct 43 VFQRIALWLLSTGSAPIKLNDA-ATRAKIVKCK---ESLWKLLYYAACDFFVLQVIYHEP 98
Query 143 WWPQQLGGEGAEEQLW-QGYPMQQNSVYCHLYFYVAFGHHLASLIYILK-SPWLPDFFDR 200
W + +L+ G+P Q+ + LY+ G ++ + +L DF
Sbjct 99 W--------ARDIKLYFHGWPNQELKLSIKLYYMCQCGFYVYGVAALLAWETRRKDFAVM 150
Query 201 LLPCAAALCLIYFSYMSNFLRVGVVILFCHDICDIFTCGCKAFVDTPYHKVTIGL---FV 257
+ + L+ +SY+++F R+G +IL HD D+F K F Y + G F
Sbjct 151 MSHHVITIILLSYSYLTSFFRIGAIILALHDASDVFMETAKIF---KYSEKEFGASVCFA 207
Query 258 LLTVCWFYFRLYTFPSAALFPIYKAIKTRPDHSETEGS-SYFIF--ILLTLSLMNIYWFV 314
L V W RL FP + + D + EG+ Y+ F +LL L + +IYW+
Sbjct 208 LFAVSWLLLRLIYFPFWIIRATSIELLDYLDMTSAEGTLMYYSFNTMLLMLLVFHIYWWY 267
Query 315 LMVKMFVHFILSGQMRDLHSRVSEEVSSAQSRSRE 349
L+ M V + + +V E++ S +
Sbjct 268 LICAMIVRLLKN------RGKVGEDIRSDSEDDDD 296
> 7290690
Length=339
Score = 55.8 bits (133), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 63/254 (24%), Positives = 106/254 (41%), Gaps = 63/254 (24%)
Query 117 FGEMTWKMLYFLMTTAVCLLSFRNERW-WPQQLGGEGAEEQLWQGYPMQQNSVYCHLYFY 175
F E TW+ +Y+L + ++ ++ W W + W GYP Q
Sbjct 110 FCENTWRCIYYLYSFIFGVIVLWDKPWFWDVK--------SCWYGYPHQM---------- 151
Query 176 VAFGHHLASLIYILKSPWLPDFFDRLLPCAAALCLIYFSYMSNFLRVGVVILFCHDICDI 235
F HH+ +L+ L+ S++ N RVG ++L HD DI
Sbjct 152 --FIHHMVTLL-----------------------LMSLSWVCNLHRVGSLVLVVHDCADI 186
Query 236 FTCGCKAFVDTPYHKVTIGLFVLLTVCWFYFRLYTFPSAALFPIYKAIKTRPDHSETEGS 295
F K Y K+ +F + TV W RL +P +A + P
Sbjct 187 FLEAAKLTKYAKYQKLCDAIFAIFTVVWIVTRLGFYPRIIYSSSVEAPRILPMF-----P 241
Query 296 SYFIF--ILLTLSLMNIYWFVLMVKMFVHFILSGQMR-DLHSRVSEEVS----------- 341
+Y+IF +LL L ++++ W +++K+ V + G M D+ S SE+++
Sbjct 242 AYYIFNSLLLMLLVLHVIWTYMILKIVVDSLQKGLMSGDIRSSDSEDLTDSSGNARLTNG 301
Query 342 SAQSRSREISSKAS 355
SA+S+++ ISS S
Sbjct 302 SARSKNKSISSAPS 315
> Hs20532972
Length=252
Score = 54.3 bits (129), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 32/136 (23%), Positives = 63/136 (46%), Gaps = 11/136 (8%)
Query 103 DRW----SEQVRQLLCSHFGEMTWKMLYFLMTTAVCLLSFRNERWWPQQLGGEGAEEQLW 158
+RW Q R L F E +W+ ++L+ + ++ W+ +++W
Sbjct 115 ERWFRRRRNQDRPSLLKKFREASWRFTFYLIAFIAGMAVIVDKPWFYDM-------KKVW 167
Query 159 QGYPMQQNSVYCHLYFYVAFGHHLASLIYILKSPWLPDFFDRLLPCAAALCLIYFSYMSN 218
+GYP+Q + Y+ + + + L I DF ++++ A + LI FS+ +N
Sbjct 168 EGYPIQSTIPSQYWYYMIELSFYWSLLFSIASDVKRKDFKEQIIHHVATIILISFSWFAN 227
Query 219 FLRVGVVILFCHDICD 234
++R G +I+ HD D
Sbjct 228 YIRAGTLIMALHDSSD 243
> Hs11641421
Length=350
Score = 53.9 bits (128), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 62/193 (32%), Positives = 92/193 (47%), Gaps = 26/193 (13%)
Query 178 FGHHLASLIYILKSPWLPDFFDRLLPCAAALCLIYFSYMSNFLRVGVVILFCHDICDIFT 237
+GH + + +Y+ W D LL L LI SY + VG+++LF HDI D+
Sbjct 159 YGHSIYATLYM--DTWRKDSVVMLLHHVVTLILIVSSYAFRYHNVGILVLFLHDISDVQL 216
Query 238 CGCKAFV-----DTPYHKVTIGLFVL----LTVCWFYFRLYTFPSAALFPI-YKAIKTRP 287
K + YH++ L WF+FRLY FP L+ + +++T P
Sbjct 217 EFTKLNIYFKSRGGSYHRLHALAADLGCLSFGFSWFWFRLYWFPLKVLYATSHCSLRTVP 276
Query 288 DHSETEGSSYFIF--ILLTLSLMNIYWFVLMVKMFVHFILSGQMRDLHSRVSEEVSSAQS 345
D YF F +LL L+LMN+YWF+ +V F +L+GQ+ +L + + AQS
Sbjct 277 D-----IPFYFFFNALLLLLTLMNLYWFLYIVA-FAAKVLTGQVHELKDLREYDTAEAQS 330
Query 346 RSREISSKASKAE 358
K SKAE
Sbjct 331 L------KPSKAE 337
> Hs20481895
Length=293
Score = 52.4 bits (124), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 51/199 (25%), Positives = 91/199 (45%), Gaps = 15/199 (7%)
Query 156 QLWQGYPMQQNSVYCHLYFYVAFGHHLASLIYILKSPWLPDFFDRLLPCAAALCLIYFSY 215
++W GYP Q + Y+ + + + L + DF ++ AA+ L+ FS+
Sbjct 78 EVWNGYPKQPLLPSQYWYYILEMSFYWSLLFRLGFDVKRKDFLAHIIHHLAAISLMSFSW 137
Query 216 MSNFLRVGVVILFCHDICDIFTCGCKAFVDTPYHKVTIGLFVLLTVCWFYFRLYTFPSAA 275
+N++R G +++ HD+ DI+ K F + + LF + + +F RL FP
Sbjct 138 CANYIRSGTLVMIVHDVADIWLESAKMFSYAGWTQTCNTLFFIFSTIFFISRLIVFPFWI 197
Query 276 LF-----PIYKAIKTRPDHSETEGSSYFIFI-LLTLSLMNIYWFVLMVKMFVHFI-LSGQ 328
L+ P+Y H E S F+ + L+ L ++++YW ++KM I +
Sbjct 198 LYCTLILPMY--------HLEPFFSYIFLNLQLMILQVLHLYWGYYILKMLNRCIFMKVT 249
Query 329 MRDLHSRVSEEVSSAQSRS 347
L +RVS E + RS
Sbjct 250 GVGLPTRVSSESGAWPQRS 268
> CE20184
Length=360
Score = 50.1 bits (118), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 39/136 (28%), Positives = 63/136 (46%), Gaps = 19/136 (13%)
Query 207 ALCLIYFSYMSNFLRVGVVILFCHDICDI------FTCGCKAFVDTPYHK----VTIGLF 256
AL L++ SY+ NF G ++LF HD D + K + Y+K + F
Sbjct 194 ALGLLFLSYVDNFTLPGALVLFLHDNSDATLEITKLSFYLKKRTNRQYYKYYFLMGNAAF 253
Query 257 VLLTVCWFYFRLYTFPSAALFP-IYKAIKTRPDHSETEGSSYFIFI---LLTLSLMNIYW 312
+L + W FRLY + L+ IY A+ P + + +F + LL + MN+YW
Sbjct 254 ILFAIIWVIFRLYWYTCKLLYATIYGAVYLGP-----QDAPFFPLLGAMLLIIFAMNVYW 308
Query 313 FVLMVKMFVHFILSGQ 328
F + +M L+G+
Sbjct 309 FNFIARMIWRVALTGE 324
> ECU08g1350
Length=287
Score = 49.7 bits (117), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 58/275 (21%), Positives = 109/275 (39%), Gaps = 21/275 (7%)
Query 79 VIYLLFQGLCTWILKPLADRLLPR-----DRWSEQVRQLLCSHFGEMTWKMLYFLMTTAV 133
V LL + I+ P A L+ R D SE+ ++ S WK +++ T+
Sbjct 29 VFSLLITMVKNLIVLPTASALIKRLGLEKDLGSEKKKKFCIS-----LWKAMFYSFTSVY 83
Query 134 CLLSFRNERWWPQQLGGEGAEEQLWQGYPMQQNSVYCHLYFYVAFGHHLASLIYILKSPW 193
R+E P+ + + W + ++ ++++ F ++ L Y+
Sbjct 84 GYFVIRSE---PRAYTAKNLMDT-WGVHGAPSKVLF---FYHLEFSYYFVELFYLFSEHA 136
Query 194 LPDFFDRLLPCAAALCLIYFSYMSNFLRVGVVILFCHDICDIFTCGCKAFVDTPYHKVTI 253
DF + + L++ SY ++ LR GV I+ HDI D F K +
Sbjct 137 YKDFLQMVTHHIVTMLLLFLSYHNDLLRAGVAIIVIHDISDPFLEIGKLTNYIHDKSLAT 196
Query 254 GLFVLLTVCWFYFRLYTFPSAALFPIYKAIKTRPDHSETEGSSYFIFILLTLSLMNIYWF 313
+F + RL + PI + + +H + +L L M+I W
Sbjct 197 SIFTCFAGIFIASRLGIYAFLLSLPI---VVSMWEHGFSPSLFLIAMLLQGLQAMHIVWS 253
Query 314 VLMVKMFVHFILSGQMRDLHSRVSEEVSSAQSRSR 348
+++V+M I ++ D+ S + E S +Q R +
Sbjct 254 LMIVRMARKVIHETELEDIRS-IKTETSPSQLRCK 287
> Hs20550645
Length=328
Score = 48.1 bits (113), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 30/118 (25%), Positives = 50/118 (42%), Gaps = 0/118 (0%)
Query 155 EQLWQGYPMQQNSVYCHLYFYVAFGHHLASLIYILKSPWLPDFFDRLLPCAAALCLIYFS 214
Q W YP Q S + Y+ + + + + DF + + LI FS
Sbjct 91 RQCWHNYPFQPLSSGLYHYYIMELAFYWSLMFSQFTDIKRKDFLIMFVHHLVTIGLISFS 150
Query 215 YMSNFLRVGVVILFCHDICDIFTCGCKAFVDTPYHKVTIGLFVLLTVCWFYFRLYTFP 272
Y++N +RVG +I+ HD+ D K Y ++ LFV+ + + RL +P
Sbjct 151 YINNMVRVGTLIMCLHDVSDFLLEAAKLANYAKYQRLCDTLFVIFSAVFMVTRLGIYP 208
> YHL003c
Length=411
Score = 45.8 bits (107), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 44/207 (21%), Positives = 89/207 (42%), Gaps = 19/207 (9%)
Query 157 LWQGYPMQQNSVYCHLYFY--VAF-GHHLASLIYILKSPWLPDFFDRLLPCAAALCLIYF 213
+++ YP+ N +++ AF L+ L+ P D+ + + L LI+
Sbjct 207 MYRTYPVITNPFLFKIFYLGQAAFWAQQACVLVLQLEKP-RKDYKELVFHHIVTLLLIWS 265
Query 214 SYMSNFLRVGVVILFCHDICDIFTCGCKA--FVDTPYHKVTIGLFVLLTVCWFYFRLYTF 271
SY+ +F ++G+ I D+ D F K ++++ + GLFV W Y R +
Sbjct 266 SYVFHFTKMGLAIYITMDVSDFFLSLSKTLNYLNSVFTPFVFGLFVFF---WIYLR-HVV 321
Query 272 PSAALFPIYKAIKTRPDHSETEGSSYF--------IFILLT-LSLMNIYWFVLMVKMFVH 322
L+ + + ++ + + +F+L+ L L+N+YW L++++
Sbjct 322 NIRILWSVLTEFRHEGNYVLNFATQQYKCWISLPIVFVLIAALQLVNLYWLFLILRILYR 381
Query 323 FILSGQMRDLHSRVSEEVSSAQSRSRE 349
I G +D S + S+ S+E
Sbjct 382 LIWQGIQKDERSDSDSDESAENEESKE 408
> Hs22044511
Length=192
Score = 40.4 bits (93), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 40/140 (28%), Positives = 65/140 (46%), Gaps = 14/140 (10%)
Query 196 DFFDRLLPCAAALCLIYFSYMSNFLRVGVVILFCHDICDIFTCGCKAFVDTPYHKVTIGL 255
DF L ++ LI FSY++N RVG ++L HD D K + K+ L
Sbjct 12 DFGIMFLHHLVSIFLITFSYVNNMARVGTLVLCLHDSADALLEAAKMANYAKFQKMCDLL 71
Query 256 FVLLTVCWFYFRLYTFP----SAALFPIYKAIKTRPDHSETEGSSYFIFILLTLSL--MN 309
FV+ V + RL FP + LF ++ + P S+++F LL L + +N
Sbjct 72 FVMFAVVFITTRLGIFPLWVLNTTLFESWEIVGPYP--------SWWVFNLLLLLVQGLN 123
Query 310 IYWFVLMVKMFVHFILSGQM 329
+W L+VK+ + G++
Sbjct 124 CFWSYLIVKIACKAVSRGKV 143
> SPBC3E7.15c
Length=384
Score = 38.5 bits (88), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 74/336 (22%), Positives = 138/336 (41%), Gaps = 48/336 (14%)
Query 10 ESASQVSRCTCNSKMVRLDRVFG---ILLAVIAGWSAFHYPKVAGSIRHRLSVPGGGYPD 66
S S V R N+ + ++ + ILL ++ GW ++ G I++ + + YP
Sbjct 38 RSKSIVGRAAQNAVLRSKEKTWIVPLILLTLLVGW---YFVNPNGYIKYGIFL---SYPI 91
Query 67 P-SDLRIYGS-----AAAVIYLLFQGLC-TWILKPLADRLLP--RDRWSEQVRQLLCSHF 117
P ++ YG A + Y LF C +I++ + R+ R ++R+ F
Sbjct 92 PGTNPAQYGKGRLDIAFCLFYALFFTFCREFIMQEIIARIGRHFNIRAPAKLRR-----F 146
Query 118 GEMTWKMLYFLMTTAVCLLSFRNERWWPQQLGGEGAEEQLWQGYPMQQNSVYCHLYFYVA 177
E + LYF + + L + W + W+ YP + ++ +
Sbjct 147 EEQAYTCLYFTVMGSWGLYVMKQTPMWFFN------TDAFWEEYPHFYHVGSFKAFYLIE 200
Query 178 FGHHLAS---LIYILKSPWLPDFFDRLLPCAAALCLIYFSYMSNFLRVGVVILFCHDICD 234
+ + LI L+ P DF + ++ L LI SY +F +G+ + D D
Sbjct 201 AAYWIQQALVLILQLEKP-RKDFKELVVHHIITLLLIGLSYYFHFTWIGLAVFITMDTSD 259
Query 235 IFTC--GCKAFVDTPYHKVTIGLFVLLTVCWFYFRLYTFPSAALFPIYKAIKT------- 285
I+ C +V+T + +FV+ W Y R Y ++ ++ ++T
Sbjct 260 IWLALSKCLNYVNTV---IVYPIFVIFVFVWIYMRHY-LNFKIMWAVWGTMRTINSFDLD 315
Query 286 -RPDHSETEGSSYFIFILLT-LSLMNIYWFVLMVKM 319
+ + S ILLT L L+NIYW +L++++
Sbjct 316 WAAEQYKCWISRDVTLILLTALQLVNIYWLILILRI 351
> CE04706
Length=368
Score = 36.2 bits (82), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 37/168 (22%), Positives = 69/168 (41%), Gaps = 14/168 (8%)
Query 196 DFFDRLLPCAAALCLIYFSYMSNFLRVGVVILFCHDICDIFTCGCKAFVDTPYHKVTIGL 255
DF+ L+ L LI S+ N +RVG +IL HD DI K +
Sbjct 200 DFWQMLVHHFITLALIGVSWTMNMVRVGTLILVSHDAVDILIDVGKILRYEQFETALTIC 259
Query 256 FVLLTVCWFYFRLYTFP----SAALFPIYKAIKTRPD----HSETEGSSYFIFILLTLSL 307
F + W RL +P + F I+ + + + + + +L L +
Sbjct 260 FAGVLFVWVATRLVYYPFWIIRSVWFDAPALIQDDYEWLNFDQQPQAPRFIMLLLTALLI 319
Query 308 MNIYWFVLMVKMFVHFILSGQMRDL------HSRVSEEVSSAQSRSRE 349
++I+W ++ K+ I G + D+ S V+ E + Q+++++
Sbjct 320 LHIFWAYILFKIAYDTIQEGVVDDVREDFDEQSLVNREKAKQQNKNKD 367
> 7290103
Length=368
Score = 34.3 bits (77), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 22/88 (25%), Positives = 40/88 (45%), Gaps = 2/88 (2%)
Query 152 GAEEQLWQGYPMQQNSVYCHLYFYVAFGHHLASL--IYILKSPWLPDFFDRLLPCAAALC 209
G QLW+G+P S YF V ++L L +Y K + +++ +
Sbjct 153 GQVAQLWEGFPDHPMSFLHKFYFVVQLAYYLHMLPELYFQKIKTKEEQQPKIVHSISGFT 212
Query 210 LIYFSYMSNFLRVGVVILFCHDICDIFT 237
LI +Y +F R+ +V+L H ++ +
Sbjct 213 LIVLAYTLSFQRLALVLLTLHYFSELLS 240
> YKL008c
Length=418
Score = 33.1 bits (74), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 30/136 (22%), Positives = 60/136 (44%), Gaps = 15/136 (11%)
Query 206 AALCLIYFSYMSNFLRVGVVILFCHDICDIFTCGCKA--FVDTPYHKVTIGLFVLLTVCW 263
L LI+ SY+ +F ++G+ I D+ D K ++D+ + +FV V W
Sbjct 258 VTLLLIWSSYVFHFTKMGLPIYITMDVSDFLLSFSKTLNYLDSGLAFFSFAIFV---VAW 314
Query 264 FYFRLYTFPSAALFPIYKAIKTRPDHSETEGSSYF--------IFILL-TLSLMNIYWFV 314
Y R Y L+ + +T ++ + + +F+L+ L L+N+YW
Sbjct 315 IYLRHYI-NLKILWSVLTQFRTEGNYVLNFATQQYKCWISLPIVFVLIGALQLVNLYWLF 373
Query 315 LMVKMFVHFILSGQMR 330
L+ ++ + G ++
Sbjct 374 LIFRVLYRILWRGILK 389
> At5g48120
Length=1152
Score = 31.6 bits (70), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 23/40 (57%), Gaps = 1/40 (2%)
Query 88 CTWILKPLADRLLPRDRWSEQVRQLLCSHFGEMTWKMLYF 127
C+ +LK L ++LLPR + + QLL HF WK + F
Sbjct 672 CSQLLKCLTNKLLPRVAEIDGLEQLLV-HFAISMWKQIEF 710
> CE09961
Length=301
Score = 31.2 bits (69), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 32/57 (56%), Gaps = 4/57 (7%)
Query 251 VTIGLFVLLTVCWFYFRLYTFPSAALFPI-YKAIKTRPDHSETEGSSYFIFILLTLS 306
+ I L +LL +C+F ++ + + PI Y++++T T+ FIF+ LTLS
Sbjct 150 IDISLVILLIICFFGYKAFNLAYESAAPIHYRSLETT---KVTKAQKVFIFLFLTLS 203
Lambda K H
0.329 0.140 0.466
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8699662928
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40