bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
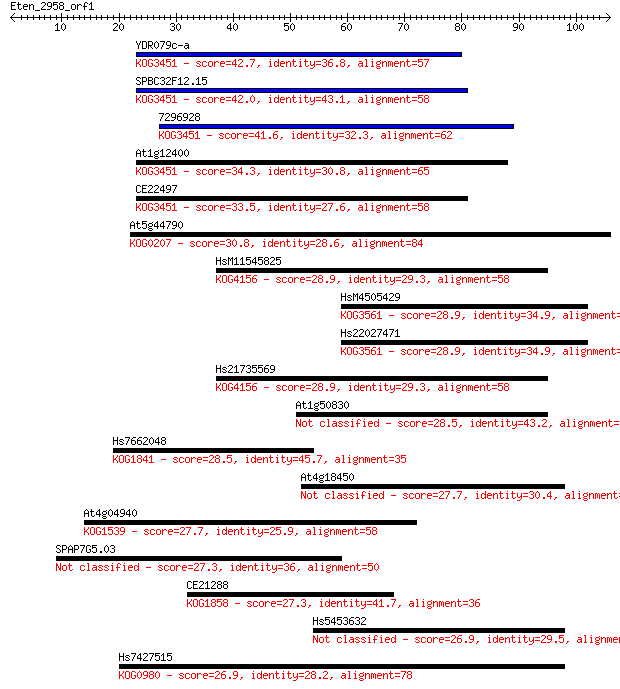
Query= Eten_2958_orf1
Length=105
Score E
Sequences producing significant alignments: (Bits) Value
YDR079c-a 42.7 2e-04
SPBC32F12.15 42.0 3e-04
7296928 41.6 3e-04
At1g12400 34.3 0.057
CE22497 33.5 0.093
At5g44790 30.8 0.65
HsM11545825 28.9 2.6
HsM4505429 28.9 2.7
Hs22027471 28.9 2.7
Hs21735569 28.9 2.7
At1g50830 28.5 3.1
Hs7662048 28.5 3.5
At4g18450 27.7 4.8
At4g04940 27.7 5.8
SPAP7G5.03 27.3 6.1
CE21288 27.3 6.8
Hs5453632 26.9 9.2
Hs7427515 26.9 9.4
> YDR079c-a
Length=72
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 36/58 (62%), Gaps = 1/58 (1%)
Query 23 MVVAIKGILVQCDPPTMEIILQLN-ETKNFIIEKISEEHVLCSESVCSFLEEEVTRRL 79
M A KG LVQCDP +ILQ++ + + ++E++ + H+L + S F++ E+ R L
Sbjct 1 MARARKGALVQCDPSIKALILQIDAKMSDIVLEELDDTHLLVNPSKVEFVKHELNRLL 58
> SPBC32F12.15
Length=68
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/60 (41%), Positives = 39/60 (65%), Gaps = 2/60 (3%)
Query 23 MVVAIKGIL-VQCDPPTMEIILQLNE-TKNFIIEKISEEHVLCSESVCSFLEEEVTRRLE 80
M A KG+L V+CDP ++IL ++E + +IE+I EE +L +ES ++ E+ RRLE
Sbjct 1 MPRAQKGLLLVECDPTVKQLILNMDEQSPGIVIEEIDEERLLVNESRLEQVKAELERRLE 60
> 7296928
Length=69
Score = 41.6 bits (96), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 35/66 (53%), Gaps = 4/66 (6%)
Query 27 IKGILVQCDPPTMEIILQLNET----KNFIIEKISEEHVLCSESVCSFLEEEVTRRLELA 82
+KG+LV+CDP + +L L+E + FII+ + E H+ S + L+ V ++
Sbjct 1 MKGVLVECDPAMKQFLLHLDEKLALGRKFIIQDLDENHLFISTDIVEVLQARVDDLMDRI 60
Query 83 ERPLAE 88
PL +
Sbjct 61 SFPLHD 66
> At1g12400
Length=71
Score = 34.3 bits (77), Expect = 0.057, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 33/71 (46%), Gaps = 6/71 (8%)
Query 23 MVVAIKGILVQCDPPTMEIILQLNE----TKNFIIEKISEEHVLCSESVCSFLEEEVT-- 76
MV AIKG+ V CD P + I+ +N ++ FII + H+ V + ++
Sbjct 1 MVNAIKGVFVSCDIPMTQFIVNMNNSMPPSQKFIIHVLDSTHLFVQPHVEQMIRSAISDF 60
Query 77 RRLELAERPLA 87
R E+P +
Sbjct 61 RDQNSYEKPTS 71
> CE22497
Length=71
Score = 33.5 bits (75), Expect = 0.093, Method: Compositional matrix adjust.
Identities = 16/62 (25%), Positives = 35/62 (56%), Gaps = 4/62 (6%)
Query 23 MVVAIKGILVQCDPPTMEIILQLNETKN----FIIEKISEEHVLCSESVCSFLEEEVTRR 78
MV KG+LV DP ++++ L++++ FI+ ++ + H+ + + LE +V +
Sbjct 1 MVNVKKGVLVTSDPAFRQLLIHLDDSRQLGSKFIVRELDDTHLFIEKEIVPMLENKVEQI 60
Query 79 LE 80
+E
Sbjct 61 ME 62
> At5g44790
Length=1001
Score = 30.8 bits (68), Expect = 0.65, Method: Composition-based stats.
Identities = 24/92 (26%), Positives = 49/92 (53%), Gaps = 11/92 (11%)
Query 22 KMVVAIKGILVQCDPPTME-IILQLNETKNFIIEKISEE-------HVLCSESVCSFLEE 73
K+V+ + GIL + D +E I+ +LN + F +++IS E V+ S S+ +EE
Sbjct 208 KLVLRVDGILNELDAQVLEGILTRLNGVRQFRLDRISGELEVVFDPEVVSSRSLVDGIEE 267
Query 74 EVTRRLELAERPLAEMQKEKKANEAGAAANTY 105
+ + +L R ++ ++ + + G A+N +
Sbjct 268 DGFGKFKL--RVMSPYER-LSSKDTGEASNMF 296
> HsM11545825
Length=1332
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 17/59 (28%), Positives = 32/59 (54%), Gaps = 1/59 (1%)
Query 37 PTMEIILQLNETKNFIIE-KISEEHVLCSESVCSFLEEEVTRRLELAERPLAEMQKEKK 94
P +E+ LQ + +F + K S++H+ E + + RRLE ER + ++++ KK
Sbjct 130 PCLELSLQSGNSTDFTTDRKSSKKHIHDKEGTAGKAKVKSKRRLEKEERKMEKIRQLKK 188
> HsM4505429
Length=824
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 24/44 (54%), Gaps = 1/44 (2%)
Query 59 EHVLCSESVCSFLEEEVTRRLELA-ERPLAEMQKEKKANEAGAA 101
E ++C+ SV S+ + V RR ELA E P +E + G++
Sbjct 342 EFIVCTHSVVSYADVRVERRQELALEDPPSEALHSSALKDKGSS 385
> Hs22027471
Length=824
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 24/44 (54%), Gaps = 1/44 (2%)
Query 59 EHVLCSESVCSFLEEEVTRRLELA-ERPLAEMQKEKKANEAGAA 101
E ++C+ SV S+ + V RR ELA E P +E + G++
Sbjct 342 EFIVCTHSVVSYADVRVERRQELALEDPPSEALHSSALKDKGSS 385
> Hs21735569
Length=1339
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 17/59 (28%), Positives = 32/59 (54%), Gaps = 1/59 (1%)
Query 37 PTMEIILQLNETKNFIIE-KISEEHVLCSESVCSFLEEEVTRRLELAERPLAEMQKEKK 94
P +E+ LQ + +F + K S++H+ E + + RRLE ER + ++++ KK
Sbjct 130 PCLELSLQSGNSTDFTTDRKSSKKHIHDKEGTAGKAKVKSKRRLEKEERKMEKIRQLKK 188
> At1g50830
Length=768
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 19/46 (41%), Positives = 29/46 (63%), Gaps = 5/46 (10%)
Query 51 FIIEKISEEHVLCSESVCSFL--EEEVTRRLELAERPLAEMQKEKK 94
F++ K SE+H C+E +CS + +EE+ RL+ ER LA + E K
Sbjct 695 FLVHKNSEKH-RCNEKMCSEVKKQEEIDERLK--ERKLAIKEMELK 737
> Hs7662048
Length=1539
Score = 28.5 bits (62), Expect = 3.5, Method: Composition-based stats.
Identities = 16/36 (44%), Positives = 21/36 (58%), Gaps = 1/36 (2%)
Query 19 FLAKMVVAIKGILVQCDPPTME-IILQLNETKNFII 53
FLAK + G++VQ P TM + L L E K+F I
Sbjct 1333 FLAKSSIVEDGLMVQITPETMNGLRLALREQKDFKI 1368
> At4g18450
Length=303
Score = 27.7 bits (60), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/46 (30%), Positives = 26/46 (56%), Gaps = 2/46 (4%)
Query 52 IIEKISEEHVLCSESVCSFLEEEVTRRLELAERPLAEMQKEKKANE 97
+++ + E H C+ S CS E TR L +E +++K +++NE
Sbjct 217 VVDGMVENH--CALSYCSTKEHSETRGLRGSEETWFDLRKRRRSNE 260
> At4g04940
Length=931
Score = 27.7 bits (60), Expect = 5.8, Method: Composition-based stats.
Identities = 15/58 (25%), Positives = 31/58 (53%), Gaps = 1/58 (1%)
Query 14 FKFGDFLAKMVVAIKGILVQCDPPTMEIILQLNETKNFIIEKISEEHVLCSESVCSFL 71
F+F + K+ + I G ++C P E +L ET++ + +K+ E+ + + +FL
Sbjct 870 FEFMQAVVKLFLKIHGETIRCHPSLQEKAKKLLETQSLVWQKM-EKLFQSTRCIVTFL 926
> SPAP7G5.03
Length=703
Score = 27.3 bits (59), Expect = 6.1, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 28/51 (54%), Gaps = 1/51 (1%)
Query 9 FFFFI-FKFGDFLAKMVVAIKGILVQCDPPTMEIILQLNETKNFIIEKISE 58
F FFI F +G +L + +AI GIL EI +N+T + I ++I +
Sbjct 104 FIFFIEFSYGTYLCLIQLAIDGILDAVADVGEEIGTAVNDTLHAIADEIED 154
> CE21288
Length=1461
Score = 27.3 bits (59), Expect = 6.8, Method: Composition-based stats.
Identities = 15/36 (41%), Positives = 17/36 (47%), Gaps = 0/36 (0%)
Query 32 VQCDPPTMEIILQLNETKNFIIEKISEEHVLCSESV 67
V CDPPT EI E NF S + SE+V
Sbjct 686 VTCDPPTTEIFKLCTEWGNFYNSLASALRIGSSETV 721
> Hs5453632
Length=1120
Score = 26.9 bits (58), Expect = 9.2, Method: Composition-based stats.
Identities = 13/46 (28%), Positives = 25/46 (54%), Gaps = 2/46 (4%)
Query 54 EKISEEHVLCSE--SVCSFLEEEVTRRLELAERPLAEMQKEKKANE 97
E ++E ++ +E S+CS L+E + +R + E+Q +A E
Sbjct 968 ESLAEMSIMTTELQSLCSLLQESKEEAIRTLQRKICELQARLQAQE 1013
> Hs7427515
Length=914
Score = 26.9 bits (58), Expect = 9.4, Method: Composition-based stats.
Identities = 22/86 (25%), Positives = 43/86 (50%), Gaps = 8/86 (9%)
Query 20 LAKMVVAIKGILVQCDPPTMEIILQLNETKNFIIEKISEEHVLCSESV--CSFLEEEVT- 76
L + + +K L + ++LQL + + ++E+ L ++ C FL E+
Sbjct 257 LYREISGLKAQLENMKTESQRVVLQLKGHVSELEADLAEQQHLRQQAADDCEFLRAELDE 316
Query 77 -RR----LELAERPLAEMQKEKKANE 97
RR E A+R L+E++++ +ANE
Sbjct 317 LRRQREDTEKAQRSLSEIERKAQANE 342
Lambda K H
0.326 0.139 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1170944580
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40