bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2937_orf1
Length=200
Score E
Sequences producing significant alignments: (Bits) Value
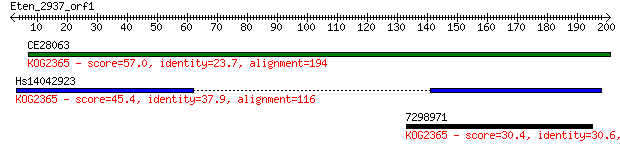
CE28063 57.0 3e-08
Hs14042923 45.4 7e-05
7298971 30.4 2.6
> CE28063
Length=808
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 46/194 (23%), Positives = 82/194 (42%), Gaps = 58/194 (29%)
Query 7 LSASVNEKIRAILFSSVKKVWFYSLFTWFIFESTGMPVVYVPTAASSLLALLPILPPEVI 66
++ ++ I + S K FY L+T+F+ + +V+VP+ A+++ A +PI+PP ++
Sbjct 644 ITVAIEHAIFGVFVLSAKMAVFYGLYTYFVHSLFDLNIVFVPSMAATVFAAIPIMPPYIV 703
Query 67 SLLPAVALWLHSDAAAAAAAAAGGGPPAEAAAAAAAAAAAAAPGGAGTPAAAAAAAAAAA 126
++ V LWL E AAA A+ AP
Sbjct 704 AIFGIVELWL---------------VRGEGAAALVFTVASFAP----------------- 731
Query 127 AGGWGHWFAAPHRLAALAVLAANALVWWNVTTCIYREIPDASPWLVGLSAALGLSTLGLK 186
V+ A+A YRE+ + P++ GL+ G+ LGL+
Sbjct 732 ------------------VMFADA--------TFYREVKGSHPYVTGLAIIGGMYWLGLQ 765
Query 187 GIILGPVLATVPLI 200
G I+GP++ + L+
Sbjct 766 GAIIGPIVLCLCLV 779
> Hs14042923
Length=911
Score = 45.4 bits (106), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 36/59 (61%), Gaps = 0/59 (0%)
Query 3 SSAILSASVNEKIRAILFSSVKKVWFYSLFTWFIFESTGMPVVYVPTAASSLLALLPIL 61
SS I+ SV E IR + +S+K FY L+TW G+ +V++P+A +++L +P L
Sbjct 695 SSNIIGQSVEEAIRGVFDASLKMAGFYGLYTWLTHTMFGINIVFIPSALAAILGAVPFL 753
Score = 33.1 bits (74), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 33/58 (56%), Gaps = 1/58 (1%)
Query 141 AALAVLAANALVWWNVTTCIYREIPDA-SPWLVGLSAALGLSTLGLKGIILGPVLATV 197
A+ +L + L + V T IY +I P+L GL+ A G LGL+G I+GP+L +
Sbjct 775 KAILLLIFHLLPTYFVDTAIYSDISGGGHPYLTGLAVAGGAYYLGLEGAIIGPILLCI 832
> 7298971
Length=837
Score = 30.4 bits (67), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 30/62 (48%), Gaps = 0/62 (0%)
Query 133 WFAAPHRLAALAVLAANALVWWNVTTCIYREIPDASPWLVGLSAALGLSTLGLKGIILGP 192
W A A L + V + T IY ++ P+L GL+ A G+ +G +G I GP
Sbjct 745 WLAQDRFYAGLILFLLQFFVPSSFETAIYADLKGGHPYLNGLAIAGGMYWIGWQGAIFGP 804
Query 193 VL 194
++
Sbjct 805 LM 806
Lambda K H
0.319 0.129 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3479363422
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40