bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2931_orf2
Length=202
Score E
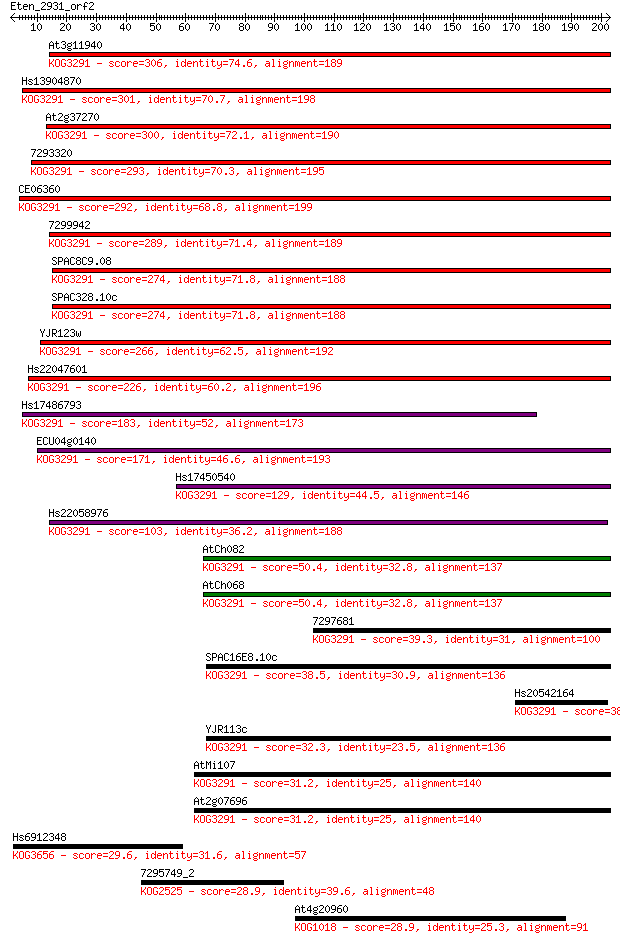
Sequences producing significant alignments: (Bits) Value
At3g11940 306 2e-83
Hs13904870 301 5e-82
At2g37270 300 1e-81
7293320 293 2e-79
CE06360 292 2e-79
7299942 289 3e-78
SPAC8C9.08 274 9e-74
SPAC328.10c 274 9e-74
YJR123w 266 2e-71
Hs22047601 226 3e-59
Hs17486793 183 2e-46
ECU04g0140 171 8e-43
Hs17450540 129 6e-30
Hs22058976 103 2e-22
AtCh082 50.4 2e-06
AtCh068 50.4 2e-06
7297681 39.3 0.006
SPAC16E8.10c 38.5 0.010
Hs20542164 38.1 0.011
YJR113c 32.3 0.59
AtMi107 31.2 1.6
At2g07696 31.2 1.6
Hs6912348 29.6 4.5
7295749_2 28.9 6.6
At4g20960 28.9 7.2
> At3g11940
Length=207
Score = 306 bits (784), Expect = 2e-83, Method: Compositional matrix adjust.
Identities = 141/190 (74%), Positives = 166/190 (87%), Gaps = 1/190 (0%)
Query 14 DFKLFGRWTYDDVTVSDLSLVDYLAVKG-KAAVLTPHTAGRYQKKRFRKAQCPLVERLVN 72
+ KLF RWTYDDVTV+D+SLVDY+ V+ K A PHTAGRY KRFRKAQCP+VERL N
Sbjct 18 EVKLFNRWTYDDVTVTDISLVDYIGVQAAKHATFVPHTAGRYSVKRFRKAQCPIVERLTN 77
Query 73 SLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIGSAGVV 132
SLMMHGRN+GKKL A+RIV+HA +II L++D NP+QV++ A+ GPRED+TRIGSAGVV
Sbjct 78 SLMMHGRNNGKKLMAVRIVKHAMEIIHLLSDLNPIQVIIDAIVNSGPREDATRIGSAGVV 137
Query 133 RRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYAIKKKD 192
RRQAVD+SPLRRVNQAI+LI TGAR AAFRN+K+++ECLADE++N AK SSNSYAIKKKD
Sbjct 138 RRQAVDISPLRRVNQAIFLITTGAREAAFRNIKTIAECLADELINAAKGSSNSYAIKKKD 197
Query 193 EIERVAKANR 202
EIERVAKANR
Sbjct 198 EIERVAKANR 207
> Hs13904870
Length=204
Score = 301 bits (771), Expect = 5e-82, Method: Compositional matrix adjust.
Identities = 140/198 (70%), Positives = 166/198 (83%), Gaps = 0/198 (0%)
Query 5 AAAATMAATDFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQC 64
AA A D KLFG+W+ DDV ++D+SL DY+AVK K A PH+AGRY KRFRKAQC
Sbjct 7 AAPAVAETPDIKLFGKWSTDDVQINDISLQDYIAVKEKYAKYLPHSAGRYAAKRFRKAQC 66
Query 65 PLVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDST 124
P+VERL NS+MMHGRN+GKKL +RIV+HAF+II L+ +NPLQVLV A+ GPREDST
Sbjct 67 PIVERLTNSMMMHGRNNGKKLMTVRIVKHAFEIIHLLTGENPLQVLVNAIINSGPREDST 126
Query 125 RIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSN 184
RIG AG VRRQAVDVSPLRRVNQAI+L+CTGAR AAFRN+K+++ECLADE++N AK SSN
Sbjct 127 RIGRAGTVRRQAVDVSPLRRVNQAIWLLCTGAREAAFRNIKTIAECLADELINAAKGSSN 186
Query 185 SYAIKKKDEIERVAKANR 202
SYAIKKKDE+ERVAK+NR
Sbjct 187 SYAIKKKDELERVAKSNR 204
> At2g37270
Length=207
Score = 300 bits (768), Expect = 1e-81, Method: Compositional matrix adjust.
Identities = 137/191 (71%), Positives = 166/191 (86%), Gaps = 1/191 (0%)
Query 13 TDFKLFGRWTYDDVTVSDLSLVDYLAVK-GKAAVLTPHTAGRYQKKRFRKAQCPLVERLV 71
+ KLF RW++DDV+V+D+SLVDY+ V+ K A PHTAGRY KRFRKAQCP+VERL
Sbjct 17 NEVKLFNRWSFDDVSVTDISLVDYIGVQPSKHATFVPHTAGRYSVKRFRKAQCPIVERLT 76
Query 72 NSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIGSAGV 131
NSLMMHGRN+GKKL A+RIV+HA +II L++D NP+QV++ A+ GPRED+TRIGSAGV
Sbjct 77 NSLMMHGRNNGKKLMAVRIVKHAMEIIHLLSDLNPIQVIIDAIVNSGPREDATRIGSAGV 136
Query 132 VRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYAIKKK 191
VRRQAVD+SPLRRVNQAI+L+ TGAR AAFRN+K+++ECLADE++N AK SSNSYAIKKK
Sbjct 137 VRRQAVDISPLRRVNQAIFLLTTGAREAAFRNIKTIAECLADELINAAKGSSNSYAIKKK 196
Query 192 DEIERVAKANR 202
DEIERVAKANR
Sbjct 197 DEIERVAKANR 207
> 7293320
Length=228
Score = 293 bits (749), Expect = 2e-79, Method: Compositional matrix adjust.
Identities = 137/195 (70%), Positives = 164/195 (84%), Gaps = 0/195 (0%)
Query 8 ATMAATDFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQCPLV 67
+T + KLFGRW+ DDVTV+D+SL DY++VK K A PH+AGRY KRFRKAQCP+V
Sbjct 34 STTELPEIKLFGRWSCDDVTVNDISLQDYISVKEKFARYLPHSAGRYAAKRFRKAQCPIV 93
Query 68 ERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIG 127
ERL SLMM GRN+GKKL A RIV+H+F+II L+ +NPLQ+LV A+ GPREDSTRIG
Sbjct 94 ERLTCSLMMKGRNNGKKLMACRIVKHSFEIIHLLTGENPLQILVSAIINSGPREDSTRIG 153
Query 128 SAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYA 187
AG VRRQAVDVSPLRRVNQAI+L+CTGAR AAFRN+K+++ECLADE++N AK SSNSYA
Sbjct 154 RAGTVRRQAVDVSPLRRVNQAIWLLCTGAREAAFRNIKTIAECLADELINAAKGSSNSYA 213
Query 188 IKKKDEIERVAKANR 202
IKKKDE+ERVAK+NR
Sbjct 214 IKKKDELERVAKSNR 228
> CE06360
Length=210
Score = 292 bits (748), Expect = 2e-79, Method: Compositional matrix adjust.
Identities = 137/199 (68%), Positives = 165/199 (82%), Gaps = 0/199 (0%)
Query 4 AAAAATMAATDFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQ 63
A AA A + LFG+W+ V VSD+SLVDY+ VK K+A PH+AGR+Q +RFRKA
Sbjct 12 ADAAPATEAPEVALFGKWSLQSVNVSDISLVDYIPVKEKSAKYLPHSAGRFQVRRFRKAA 71
Query 64 CPLVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDS 123
CP+VERL NSLMMHGRN+GKKL +RIV+HAF+II L+ +NP+QVLV AV GPREDS
Sbjct 72 CPIVERLANSLMMHGRNNGKKLMTVRIVKHAFEIIYLLTGENPVQVLVNAVINSGPREDS 131
Query 124 TRIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESS 183
TRIG AG VRRQAVDV+PLRRVNQAI+L+CTGAR AAFRN+K+++ECLADE++N AK SS
Sbjct 132 TRIGRAGTVRRQAVDVAPLRRVNQAIWLLCTGAREAAFRNVKTIAECLADELINAAKGSS 191
Query 184 NSYAIKKKDEIERVAKANR 202
NSYAIKKKDE+ERVAK+NR
Sbjct 192 NSYAIKKKDELERVAKSNR 210
> 7299942
Length=230
Score = 289 bits (739), Expect = 3e-78, Method: Compositional matrix adjust.
Identities = 135/189 (71%), Positives = 160/189 (84%), Gaps = 0/189 (0%)
Query 14 DFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQCPLVERLVNS 73
+ KLFGRW DD+++SD+SL DY+AVK K A PH+AGRY KRFRKAQCP+VERL +
Sbjct 42 EIKLFGRWACDDISISDISLQDYIAVKEKFARYLPHSAGRYAAKRFRKAQCPIVERLTSG 101
Query 74 LMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIGSAGVVR 133
LMM GR++GKKL A RIV+HAF+II L+ +NPLQV V A+ GPREDSTRIG AG VR
Sbjct 102 LMMKGRSNGKKLLACRIVKHAFEIIHLLTSENPLQVTVNAIVNSGPREDSTRIGRAGTVR 161
Query 134 RQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYAIKKKDE 193
RQAVDVSPLRRVNQAI+LICTGAR AAFRN+K+++ECLADE++N AK SSNSYAIKKKDE
Sbjct 162 RQAVDVSPLRRVNQAIWLICTGAREAAFRNIKTVAECLADELINAAKGSSNSYAIKKKDE 221
Query 194 IERVAKANR 202
+ERVAK+NR
Sbjct 222 LERVAKSNR 230
> SPAC8C9.08
Length=203
Score = 274 bits (700), Expect = 9e-74, Method: Compositional matrix adjust.
Identities = 135/188 (71%), Positives = 158/188 (84%), Gaps = 2/188 (1%)
Query 15 FKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQCPLVERLVNSL 74
KLF ++ ++ V V D+SLVDY+ + G L PHTAGR+Q KRFRKA+C +VERL NSL
Sbjct 18 IKLFNKFPFEGVEVKDISLVDYITI-GNGQPL-PHTAGRFQTKRFRKARCFIVERLTNSL 75
Query 75 MMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIGSAGVVRR 134
MM+GRN+GKKL A RIV+HAF+IIAL+ D+NPLQVLV AV GPREDSTRIGSAG VRR
Sbjct 76 MMNGRNNGKKLLATRIVKHAFEIIALLTDQNPLQVLVDAVAACGPREDSTRIGSAGTVRR 135
Query 135 QAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYAIKKKDEI 194
QAVDVSPLRRVNQA+ LI GAR AAFRN+KS+SECLA+EI+N AK SSNSYAIKKKDE+
Sbjct 136 QAVDVSPLRRVNQALALITIGAREAAFRNVKSISECLAEEIINAAKGSSNSYAIKKKDEL 195
Query 195 ERVAKANR 202
ERVAK+NR
Sbjct 196 ERVAKSNR 203
> SPAC328.10c
Length=203
Score = 274 bits (700), Expect = 9e-74, Method: Compositional matrix adjust.
Identities = 135/188 (71%), Positives = 158/188 (84%), Gaps = 2/188 (1%)
Query 15 FKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQCPLVERLVNSL 74
KLF ++ ++ V V D+SLVDY+ + G L PHTAGR+Q KRFRKA+C +VERL NSL
Sbjct 18 IKLFNKFPFEGVEVKDISLVDYITI-GNGQPL-PHTAGRFQTKRFRKARCFIVERLTNSL 75
Query 75 MMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIGSAGVVRR 134
MM+GRN+GKKL A RIV+HAF+IIAL+ D+NPLQVLV AV GPREDSTRIGSAG VRR
Sbjct 76 MMNGRNNGKKLLATRIVKHAFEIIALLTDQNPLQVLVDAVAACGPREDSTRIGSAGTVRR 135
Query 135 QAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYAIKKKDEI 194
QAVDVSPLRRVNQA+ LI GAR AAFRN+KS+SECLA+EI+N AK SSNSYAIKKKDE+
Sbjct 136 QAVDVSPLRRVNQALALITIGAREAAFRNVKSISECLAEEIINAAKGSSNSYAIKKKDEL 195
Query 195 ERVAKANR 202
ERVAK+NR
Sbjct 196 ERVAKSNR 203
> YJR123w
Length=225
Score = 266 bits (680), Expect = 2e-71, Method: Compositional matrix adjust.
Identities = 120/192 (62%), Positives = 155/192 (80%), Gaps = 2/192 (1%)
Query 11 AATDFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQCPLVERL 70
A T+ KLF +W++++V V D SLVDY+ V+ + HTAGRY KRFRKAQCP++ERL
Sbjct 36 AQTEIKLFNKWSFEEVEVKDASLVDYVQVR--QPIFVAHTAGRYANKRFRKAQCPIIERL 93
Query 71 VNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIGSAG 130
NSLMM+GRN+GKKL+A+RI++H DII ++ D+NP+QV+V A+ GPRED+TR+G G
Sbjct 94 TNSLMMNGRNNGKKLKAVRIIKHTLDIINVLTDQNPIQVVVDAITNTGPREDTTRVGGGG 153
Query 131 VVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYAIKK 190
RRQAVDVSPLRRVNQAI L+ GAR AAFRN+K+++E LA+E++N AK SS SYAIKK
Sbjct 154 AARRQAVDVSPLRRVNQAIALLTIGAREAAFRNIKTIAETLAEELINAAKGSSTSYAIKK 213
Query 191 KDEIERVAKANR 202
KDE+ERVAK+NR
Sbjct 214 KDELERVAKSNR 225
> Hs22047601
Length=337
Score = 226 bits (575), Expect = 3e-59, Method: Compositional matrix adjust.
Identities = 118/198 (59%), Positives = 142/198 (71%), Gaps = 11/198 (5%)
Query 7 AATMAATD--FKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQC 64
AA + A + KLFG+W+ DD DY AVK K A PH+A RY KRFRKAQC
Sbjct 149 AAPLVAENPGIKLFGKWSSDD---------DYTAVKEKYAKYLPHSAERYAAKRFRKAQC 199
Query 65 PLVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDST 124
P+VE L S+M HGRN+ KKL +RIV+HAF+II L+ NPLQVLV A+ GPREDST
Sbjct 200 PIVECLTYSMMTHGRNNRKKLMIMRIVKHAFEIIQLLTGDNPLQVLVNAIINSGPREDST 259
Query 125 RIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSN 184
G A +VRRQAV VSPL RVNQAI+ CTGA AAFRN+K+++E LADE+++ K SSN
Sbjct 260 LTGRARIVRRQAVAVSPLSRVNQAIWPQCTGALEAAFRNIKTIAEFLADELISAVKGSSN 319
Query 185 SYAIKKKDEIERVAKANR 202
SYA KKKDE+E VAK+NR
Sbjct 320 SYASKKKDELECVAKSNR 337
> Hs17486793
Length=156
Score = 183 bits (465), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 90/173 (52%), Positives = 111/173 (64%), Gaps = 25/173 (14%)
Query 5 AAAATMAATDFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQC 64
AA A D KLFG+W+ DDV ++D+SL DY+AV K A PH++G Y KRF KAQC
Sbjct 7 AAPAVAQTLDIKLFGKWSTDDVQINDISLQDYIAVTEKYAKYLPHSSGWYAPKRFHKAQC 66
Query 65 PLVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDST 124
P+VE L NS+MMHGRN+ KK +RIV GP E ST
Sbjct 67 PIVEPLTNSMMMHGRNNCKKFMTVRIV-------------------------NGPWEGST 101
Query 125 RIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMN 177
RIG A VRRQAVDVSPLR VNQAI+L+CTG A FRN+K+++ECLAD+++N
Sbjct 102 RIGRARTVRRQAVDVSPLRHVNQAIWLLCTGTCEAPFRNIKTIAECLADKLIN 154
> ECU04g0140
Length=208
Score = 171 bits (433), Expect = 8e-43, Method: Compositional matrix adjust.
Identities = 90/193 (46%), Positives = 127/193 (65%), Gaps = 2/193 (1%)
Query 10 MAATDFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQCPLVER 69
+ + LF +++Y++V V+++SL Y+ + + V PH A K KA+ P+ ER
Sbjct 18 LGMAEINLFEKYSYEEVKVNNVSLRPYINLSRRGIV--PHAATTITKGTTGKARIPIAER 75
Query 70 LVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIGSA 129
V SLM HGRNSGKK AI I + A II M KNPLQVLV A+ GPRED+ RIG A
Sbjct 76 FVCSLMRHGRNSGKKRLAINIFEDACFIIHSMTKKNPLQVLVDAIVNSGPREDTARIGRA 135
Query 130 GVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYAIK 189
G +RR +VDVSPL+R++ AI + G R A+FRN +++E +A+E++ + S NSYA+
Sbjct 136 GSMRRTSVDVSPLKRISIAISNLSAGIRNASFRNRITLAEAIANELIAASTNSQNSYAVN 195
Query 190 KKDEIERVAKANR 202
KK EIER+A++NR
Sbjct 196 KKKEIERIAQSNR 208
> Hs17450540
Length=121
Score = 129 bits (323), Expect = 6e-30, Method: Compositional matrix adjust.
Identities = 65/146 (44%), Positives = 88/146 (60%), Gaps = 28/146 (19%)
Query 57 KRFRKAQCPLVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVER 116
K F K QCP++E L NS+ MHG N+GKK + IV+HAF+II L+ ++N L+VLV A+
Sbjct 4 KHFHKTQCPILEHLTNSMKMHGHNNGKKPMTMHIVKHAFEIIHLLMNENTLRVLVNAIIN 63
Query 117 GGPREDSTRIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIM 176
GP EDST IG G A+F+N+K+++ECL DE++
Sbjct 64 SGPWEDSTCIG---------------------------GTCEASFQNIKTIAECLEDELI 96
Query 177 NCAKESSNSYAIKKKDEIERVAKANR 202
N + SSNSY I KKDE+E VAK+N
Sbjct 97 NATRGSSNSYVI-KKDELEHVAKSNH 121
> Hs22058976
Length=145
Score = 103 bits (258), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 68/188 (36%), Positives = 96/188 (51%), Gaps = 59/188 (31%)
Query 14 DFKLFGRWTYDDVTVSDLSLVDYLAVKGKAAVLTPHTAGRYQKKRFRKAQCPLVERLVNS 73
D K FG+W+ DD +SD+SL DY+AV K+++ K Q P
Sbjct 16 DIKFFGKWSTDDAQISDISLQDYIAV----------------KEKYTK-QEP-------- 50
Query 74 LMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRIGSAGVVR 133
SG+ QH + P EDST I A VR
Sbjct 51 ----SAGSGE-------CQH----------------------QMWPWEDSTYIEQAETVR 77
Query 134 RQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSYAIKKKDE 193
+Q VD+SPL VNQAI+L+ TG AAF+N+K+++E LAD++++ AK SSN YAI+K +
Sbjct 78 QQDVDMSPLYHVNQAIWLLYTGTSEAAFQNIKTITEYLADKLISIAKNSSNYYAIEKNN- 136
Query 194 IERVAKAN 201
+E +AK+N
Sbjct 137 LELMAKSN 144
> AtCh082
Length=155
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 45/139 (32%), Positives = 70/139 (50%), Gaps = 13/139 (9%)
Query 66 LVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGP--REDS 123
LV LVN ++ HG KK A +I+ A I + NPL VL QA+ P +
Sbjct 22 LVNMLVNRILKHG----KKSLAYQIIYRALKKIQQKTETNPLSVLRQAIRGVTPDIAVKA 77
Query 124 TRIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESS 183
R+G G + +++ + AI + +R RN M+ L+ E+++ AK S
Sbjct 78 RRVG--GSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRN---MAFKLSSELVDAAKGSG 132
Query 184 NSYAIKKKDEIERVAKANR 202
+ AI+KK+E R+A+ANR
Sbjct 133 D--AIRKKEETHRMAEANR 149
> AtCh068
Length=155
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 45/139 (32%), Positives = 70/139 (50%), Gaps = 13/139 (9%)
Query 66 LVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGP--REDS 123
LV LVN ++ HG KK A +I+ A I + NPL VL QA+ P +
Sbjct 22 LVNMLVNRILKHG----KKSLAYQIIYRALKKIQQKTETNPLSVLRQAIRGVTPDIAVKA 77
Query 124 TRIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESS 183
R+G G + +++ + AI + +R RN M+ L+ E+++ AK S
Sbjct 78 RRVG--GSTHQVPIEIGSTQGKALAIRWLLGASRKRPGRN---MAFKLSSELVDAAKGSG 132
Query 184 NSYAIKKKDEIERVAKANR 202
+ AI+KK+E R+A+ANR
Sbjct 133 D--AIRKKEETHRMAEANR 149
> 7297681
Length=218
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 31/100 (31%), Positives = 48/100 (48%), Gaps = 4/100 (4%)
Query 103 DKNPLQVLVQAVERGGPREDSTRIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFR 162
+ NP +L QAVE P T I GV + V ++ R A+ + AR +
Sbjct 115 NTNPETLLKQAVENCRPLLQVTAIKRGGVTYQVPVPITTKRSYFLAMKWLLEAAREKERK 174
Query 163 NLKSMSECLADEIMNCAKESSNSYAIKKKDEIERVAKANR 202
S+ E LA EI++ A IK+KD++ R+ ++NR
Sbjct 175 --VSLPEKLAWEILDAA--HGQGRVIKRKDDLHRLCESNR 210
> SPAC16E8.10c
Length=241
Score = 38.5 bits (88), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 42/141 (29%), Positives = 67/141 (47%), Gaps = 19/141 (13%)
Query 67 VERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRI 126
V+ LVN +M GKK +A +IV A II +NP+ VL QA+ P ++
Sbjct 108 VQHLVNLIM----RDGKKAKAEKIVATALSIIQKETGENPIDVLKQAIAEISPL---MKL 160
Query 127 GSAGVVRRQAVDVSPL-----RRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKE 181
SA + PL RR+ A+ I ++++ K +S+ + EI+ +
Sbjct 161 VSAKRFNKSVEFPMPLKERQRRRI--ALQWILGECKSSS---PKRLSDRIVKEIIAIRSK 215
Query 182 SSNSYAIKKKDEIERVAKANR 202
+SN + KKKD + R+ NR
Sbjct 216 TSNCF--KKKDHLHRMCLVNR 234
> Hs20542164
Length=263
Score = 38.1 bits (87), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 19/31 (61%), Positives = 25/31 (80%), Gaps = 0/31 (0%)
Query 171 LADEIMNCAKESSNSYAIKKKDEIERVAKAN 201
LAD++ AK SN YAIKKKD++E+VAK+N
Sbjct 232 LADQLNIFAKGFSNFYAIKKKDKLEQVAKSN 262
> YJR113c
Length=247
Score = 32.3 bits (72), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 32/136 (23%), Positives = 67/136 (49%), Gaps = 8/136 (5%)
Query 67 VERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPREDSTRI 126
++ + N +M HG KK +A I+ A ++ ++P+Q L ++++ P T+
Sbjct 114 LDHITNMIMRHG----KKEKAQTILSRALYLVYCQTRQDPIQALEKSLDELAPLM-MTKT 168
Query 127 GSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKESSNSY 186
+ GV + + V PL + + ++A R + L +E+ AK +S+++
Sbjct 169 FNTGVAKASVIPV-PLNKRQRNRIAWNWIVQSANQRVSSDFAVRLGEELTAIAKGTSSAF 227
Query 187 AIKKKDEIERVAKANR 202
+K+D+I + A A+R
Sbjct 228 --EKRDQIHKTAIAHR 241
> AtMi107
Length=148
Score = 31.2 bits (69), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 35/140 (25%), Positives = 65/140 (46%), Gaps = 7/140 (5%)
Query 63 QCPLVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPRED 122
Q L+++LVN M GK+ + IV F A +++ ++++V AVE P +
Sbjct 8 QKLLIKKLVNFRM----KEGKRTRVRAIVYQTFHRPA-RTERDVIKLMVDAVENIKPICE 62
Query 123 STRIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKES 182
++G AG + V+ R+ AI I A S+ +C EI++ ++
Sbjct 63 VAKVGVAGTIYDVPGIVARDRQQTLAIRWILEAAFKRRISYRISLEKCSFAEILDAYQKR 122
Query 183 SNSYAIKKKDEIERVAKANR 202
+ A +K++ + +A NR
Sbjct 123 GS--ARRKRENLHGLASTNR 140
> At2g07696
Length=148
Score = 31.2 bits (69), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 35/140 (25%), Positives = 65/140 (46%), Gaps = 7/140 (5%)
Query 63 QCPLVERLVNSLMMHGRNSGKKLQAIRIVQHAFDIIALMADKNPLQVLVQAVERGGPRED 122
Q L+++LVN M GK+ + IV F A +++ ++++V AVE P +
Sbjct 8 QKLLIKKLVNFRM----KEGKRTRVRAIVYQTFHRPA-RTERDVIKLMVDAVENIKPICE 62
Query 123 STRIGSAGVVRRQAVDVSPLRRVNQAIYLICTGARTAAFRNLKSMSECLADEIMNCAKES 182
++G AG + V+ R+ AI I A S+ +C EI++ ++
Sbjct 63 VAKVGVAGTIYDVPGIVARDRQQTLAIRWILEAAFKRRISYRISLEKCSFAEILDAYQKR 122
Query 183 SNSYAIKKKDEIERVAKANR 202
+ A +K++ + +A NR
Sbjct 123 GS--ARRKRENLHGLASTNR 140
> Hs6912348
Length=353
Score = 29.6 bits (65), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 32/61 (52%), Gaps = 6/61 (9%)
Query 2 DAAAAAATMAATDFKLFGRWTYDDVTVSDLSLVDYL----AVKGKAAVLTPHTAGRYQKK 57
A ++ A + + + +F WT ++ + +V YL VK K VL+PHT+G ++
Sbjct 174 SACSSLAPIYSRSYLVF--WTVSNLMAFLIMVVVYLRIYVYVKRKTNVLSPHTSGSISRR 231
Query 58 R 58
R
Sbjct 232 R 232
> 7295749_2
Length=598
Score = 28.9 bits (63), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 19/48 (39%), Positives = 28/48 (58%), Gaps = 2/48 (4%)
Query 45 VLTPHTAGRYQKKRFRKAQCPLVERLVNSLMMHGRNSGKKLQAIRIVQ 92
VL P T+GR K RK Q P++E+ + L G + K L+AI ++Q
Sbjct 10 VLEP-TSGRATTKSSRKEQDPVIEQTLECLEKSGL-TKKDLKAIPVIQ 55
> At4g20960
Length=363
Score = 28.9 bits (63), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 23/92 (25%), Positives = 43/92 (46%), Gaps = 11/92 (11%)
Query 97 IIALMADKNPLQVLVQAVERGGPREDSTRIGSAGVVRRQAVDVSPLRRVNQA-IYLICTG 155
++ M D NP+ V + +R+ AG+ +V+ +++N+ I+ + TG
Sbjct 104 VVIGMVDPNPI-VFSSGI---------SRLKDAGIDVTVSVEEELCKKMNEGFIHRMLTG 153
Query 156 ARTAAFRNLKSMSECLADEIMNCAKESSNSYA 187
A R S++ CL D+I A +S Y+
Sbjct 154 KPFLALRYSMSVNGCLLDKIGQGASDSGGYYS 185
Lambda K H
0.320 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3551102874
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40