bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2892_orf1
Length=193
Score E
Sequences producing significant alignments: (Bits) Value
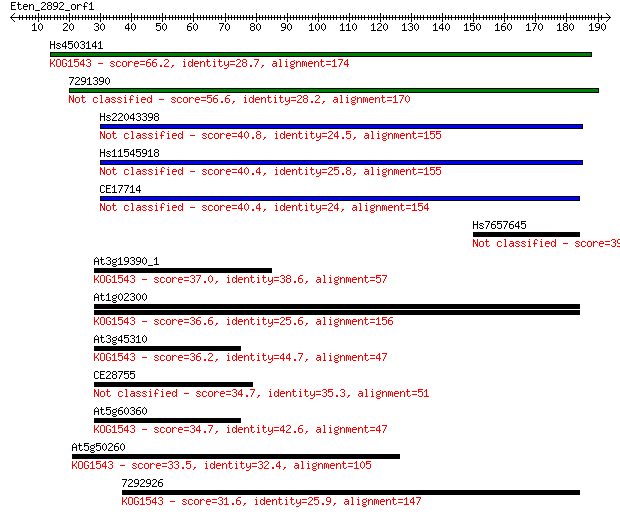
Hs4503141 66.2 4e-11
7291390 56.6 3e-08
Hs22043398 40.8 0.002
Hs11545918 40.4 0.002
CE17714 40.4 0.002
Hs7657645 39.7 0.003
At3g19390_1 37.0 0.027
At1g02300 36.6 0.029
At3g45310 36.2 0.041
CE28755 34.7 0.13
At5g60360 34.7 0.13
At5g50260 33.5 0.29
7292926 31.6 1.1
> Hs4503141
Length=463
Score = 66.2 bits (160), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 50/176 (28%), Positives = 76/176 (43%), Gaps = 53/176 (30%)
Query 14 QQQLQQLQQQQEAAVKLSPQSVLSCSFYNQGCHGGLPYLV-GKHATEIGILDEQCMPYTA 72
+ +++ L + + LSPQ V+SCS Y QGC GG PYL+ GK+A + G+++E C PYT
Sbjct 268 EARIRILTNNSQTPI-LSPQEVVSCSQYAQGCEGGFPYLIAGKYAQDFGLVEEACFPYTG 326
Query 73 MDLSSCPVLNRNKNEKDFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRW 132
D S C K ++D R+
Sbjct 327 TD-SPC-------------KMKEDCF--------------------------------RY 340
Query 133 FAKGYGYVGGCYECLSCSAEQKIMK-EIMTNGPVAAALDAPPSLFAYSSGVYTTRG 187
++ Y YVGG Y + +MK E++ +GP+A A + Y G+Y G
Sbjct 341 YSSEYHYVGGFYG----GCNEALMKLELVHHGPMAVAFEVYDDFLHYKKGIYHHTG 392
> 7291390
Length=441
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 48/174 (27%), Positives = 70/174 (40%), Gaps = 42/174 (24%)
Query 20 LQQQQEAAVKLSPQSVLSCSFYNQGCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSCP 79
+Q + + V+LS Q++LSC+ QGC GG ++ + G++DE C PYT
Sbjct 228 IQSKGKENVQLSAQNILSCTRRQQGCEGGHLDAAWRYLHKKGVVDENCYPYT-------- 279
Query 80 VLNRNKNEKDFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRWFAKGYGY 139
+D K + NG + + + L++
Sbjct 280 ------QHRDTCKIRHNSRSLRANGCQKPVNVDRDSLYT--------------------- 312
Query 140 VGGCYECLSCSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVY----TTRGAP 189
VG Y S + E IM EI +GPV A + FAYS GVY R AP
Sbjct 313 VGPAY---SLNREADIMAEIFHSGPVQATMRVNRDFFAYSGGVYRETAANRKAP 363
> Hs22043398
Length=467
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 38/160 (23%), Positives = 61/160 (38%), Gaps = 36/160 (22%)
Query 30 LSPQSVLSCSFYNQ-GCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSCPVLNRNKNEK 88
LSPQ++LSC + Q GC GG G++ + C P++ R ++E
Sbjct 254 LSPQNLLSCDTHQQQGCRGGRLDGAWWFLRRRGVVSDHCYPFSG----------RERDEA 303
Query 89 DFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRWFAKGYGYVGGCYECLS 148
G ++ + G+ T H Y Y+
Sbjct 304 ---------------GPAPPCMMHSRAMGRGKRQATAH------CPNSYVNNNDIYQVTP 342
Query 149 C----SAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVYT 184
S +++IMKE+M NGPV A ++ F Y G+Y+
Sbjct 343 VYRLGSNDKEIMKELMENGPVQALMEVHEDFFLYKGGIYS 382
> Hs11545918
Length=467
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 40/159 (25%), Positives = 64/159 (40%), Gaps = 34/159 (21%)
Query 30 LSPQSVLSCSFYNQ-GCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSCPVLNRNKNEK 88
LSPQ++LSC + Q GC GG G++ + C P++ R ++E
Sbjct 254 LSPQNLLSCDTHQQQGCRGGRLDGAWWFLRRRGVVSDHCYPFSG----------RERDEA 303
Query 89 DFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTC---HQGENRWFAKGYGYVGGCYE 145
+ + G G+ +AT C + N + Y G
Sbjct 304 G--PAPPCMMHSRAMGRGKR-----------QATAHCPNSYVNNNDIYQVTPVYRLG--- 347
Query 146 CLSCSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVYT 184
S +++IMKE+M NGPV A ++ F Y G+Y+
Sbjct 348 ----SNDKEIMKELMENGPVQALMEVHEDFFLYKGGIYS 382
> CE17714
Length=491
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 37/155 (23%), Positives = 59/155 (38%), Gaps = 35/155 (22%)
Query 30 LSPQSVLSCSFYNQ-GCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSCPVLNRNKNEK 88
LS Q +LSC+ + Q GC GG + ++G++ + C PY V +++
Sbjct 274 LSSQQLLSCNQHRQKGCEGGYLDRAWWYIRKLGVVGDHCYPY---------VSGQSREPG 324
Query 89 DFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRWFAKGYGYVGGCYECLS 148
L ++D+ + S + AF + +
Sbjct 325 HCLIPKRDYTNRQGLRCPSGSQDSTAFKMTPPYKVS------------------------ 360
Query 149 CSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVY 183
S E+ I E+MTNGPV A F Y+ GVY
Sbjct 361 -SREEDIQTELMTNGPVQATFVVHEDFFMYAGGVY 394
> Hs7657645
Length=476
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 17/34 (50%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 150 SAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVY 183
S E +IMKEIM NGPV A + F Y +G+Y
Sbjct 359 SNETEIMKEIMQNGPVQAIMQVHEDFFHYKTGIY 392
> At3g19390_1
Length=362
Score = 37.0 bits (84), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 31/59 (52%), Gaps = 2/59 (3%)
Query 28 VKLSPQSVLSC-SFYNQGCHGGLPYLVGKHATEIGILD-EQCMPYTAMDLSSCPVLNRN 84
+ LS Q ++ C + YN GC GGL K E G +D E+ PY A D++ C +N
Sbjct 174 ISLSEQELVDCDTSYNDGCGGGLMDYAFKFIIENGGIDTEEDYPYIATDVNVCNSDKKN 232
> At1g02300
Length=677
Score = 36.6 bits (83), Expect = 0.029, Method: Composition-based stats.
Identities = 40/159 (25%), Positives = 56/159 (35%), Gaps = 38/159 (23%)
Query 28 VKLSPQSVLSCSFY--NQGCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSCPVLNRNK 85
V LS +L+C + QGC+GG P ++ G++ E+C PY D + C
Sbjct 470 VSLSVNDLLACCGFLCGQGCNGGYPIAAWRYFKHHGVVTEECDPY--FDNTGC------- 520
Query 86 NEKDFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRWF-AKGYGYVGGCY 144
A+ + + C G W +K YG Y
Sbjct 521 ---------------------SHPGCEPAYP-TPKCARKCVSGNQLWRESKHYGV--SAY 556
Query 145 ECLSCSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVY 183
+ S IM E+ NGPV A Y SGVY
Sbjct 557 KVR--SHPDDIMAEVYKNGPVEVAFTVYEDFAHYKSGVY 593
Score = 30.0 bits (66), Expect = 2.6, Method: Composition-based stats.
Identities = 41/164 (25%), Positives = 56/164 (34%), Gaps = 48/164 (29%)
Query 28 VKLSPQSVLSCS--FYNQGCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSC------P 79
V LS V++C GC+GG P + G++ ++C PY D + C P
Sbjct 172 VSLSANDVIACCGLLCGFGCNGGFPMGAWLYFKYHGVVTQECDPY--FDNTGCSHPGCEP 229
Query 80 VLNRNKNEKDFLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRWFAKGYGY 139
K E+ + ++ L GE +K YG
Sbjct 230 TYPTPKCERKCV--SRNQLWGE--------------------------------SKHYGV 255
Query 140 VGGCYECLSCSAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVY 183
G Y Q IM E+ NGPV A Y SGVY
Sbjct 256 --GAYRI--NPDPQDIMAEVYKNGPVEVAFTVYEDFAHYKSGVY 295
> At3g45310
Length=377
Score = 36.2 bits (82), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 21/50 (42%), Positives = 28/50 (56%), Gaps = 3/50 (6%)
Query 28 VKLSPQSVLSC--SFYNQGCHGGLPYLVGKHATEIGILD-EQCMPYTAMD 74
+ LS Q ++ C +F N GCHGGLP ++ G LD E+ PYT D
Sbjct 186 ISLSEQQLVDCAGTFNNFGCHGGLPSQAFEYIKYNGGLDTEEAYPYTGKD 235
> CE28755
Length=383
Score = 34.7 bits (78), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 24/51 (47%), Gaps = 0/51 (0%)
Query 28 VKLSPQSVLSCSFYNQGCHGGLPYLVGKHATEIGILDEQCMPYTAMDLSSC 78
V LS Q ++ C N GC GG K E G+ E+ PY+A+ C
Sbjct 213 VSLSEQEMVDCDGRNNGCSGGYRPYAMKFVKENGLESEKEYPYSALKHDQC 263
> At5g60360
Length=358
Score = 34.7 bits (78), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 28/50 (56%), Gaps = 3/50 (6%)
Query 28 VKLSPQSVLSCS--FYNQGCHGGLPYLVGKHATEIGILD-EQCMPYTAMD 74
+ LS Q ++ C+ F N GC+GGLP ++ G LD E+ PYT D
Sbjct 186 ISLSEQQLVDCAGAFNNYGCNGGLPSQAFEYIKSNGGLDTEKAYPYTGKD 235
> At5g50260
Length=361
Score = 33.5 bits (75), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 34/117 (29%), Positives = 52/117 (44%), Gaps = 12/117 (10%)
Query 21 QQQQEAAVKLSPQSVLSC-SFYNQGCHGGLPYLVGKHATEI-GILDEQCMPYTAMDLS-- 76
Q + + LS Q ++ C + NQGC+GGL L + E G+ E PY A D +
Sbjct 164 QIRTKKLTSLSEQELVDCDTNQNQGCNGGLMDLAFEFIKEKGGLTSELVYPYKASDETCD 223
Query 77 ----SCPVLNRNKNEKDFLKSEKDFLKGEIN----GSGEDSSSNAAFLFSGEATGTC 125
+ PV++ + +E SE D +K N + + S+ F G TG C
Sbjct 224 TNKENAPVVSIDGHEDVPKNSEDDLMKAVANQPVSVAIDAGGSDFQFYSEGVFTGRC 280
> 7292926
Length=340
Score = 31.6 bits (70), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 38/154 (24%), Positives = 56/154 (36%), Gaps = 38/154 (24%)
Query 37 SCSFYNQGCHGGLPYLVGKHATEIGIL-------DEQCMPYTAMDLSSCPVLNRNKNEKD 89
+C F GC+GG P + T GI+ ++ C PY ++S C
Sbjct 151 TCGF---GCNGGFPGAAWSYWTRKGIVSGGPYGSNQGCRPY---EISPC----------- 193
Query 90 FLKSEKDFLKGEINGSGEDSSSNAAFLFSGEATGTCHQGENRWFAKGYGYVGGCYECLSC 149
+ +NG+ + + + + C G +AK + Y
Sbjct 194 ---------EHHVNGTRPPCAHGGR---TPKCSHVCQSGYTVDYAKDKHFGSKSYSVRRN 241
Query 150 SAEQKIMKEIMTNGPVAAALDAPPSLFAYSSGVY 183
E I +EIMTNGPV A L Y GVY
Sbjct 242 VRE--IQEEIMTNGPVEGAFTVYEDLILYKDGVY 273
Lambda K H
0.314 0.130 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3228275340
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40