bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2875_orf2
Length=173
Score E
Sequences producing significant alignments: (Bits) Value
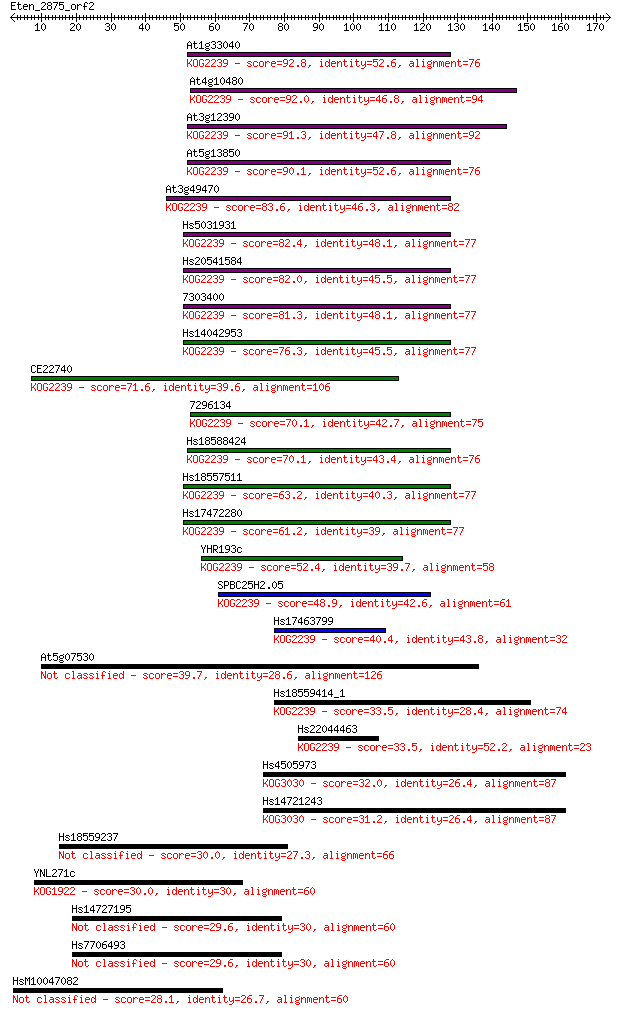
At1g33040 92.8 3e-19
At4g10480 92.0 5e-19
At3g12390 91.3 8e-19
At5g13850 90.1 2e-18
At3g49470 83.6 2e-16
Hs5031931 82.4 5e-16
Hs20541584 82.0 5e-16
7303400 81.3 1e-15
Hs14042953 76.3 3e-14
CE22740 71.6 8e-13
7296134 70.1 2e-12
Hs18588424 70.1 2e-12
Hs18557511 63.2 3e-10
Hs17472280 61.2 9e-10
YHR193c 52.4 5e-07
SPBC25H2.05 48.9 5e-06
Hs17463799 40.4 0.002
At5g07530 39.7 0.003
Hs18559414_1 33.5 0.24
Hs22044463 33.5 0.24
Hs4505973 32.0 0.61
Hs14721243 31.2 0.99
Hs18559237 30.0 2.2
YNL271c 30.0 2.6
Hs14727195 29.6 3.0
Hs7706493 29.6 3.1
HsM10047082 28.1 8.4
> At1g33040
Length=209
Score = 92.8 bits (229), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 40/78 (51%), Positives = 62/78 (79%), Gaps = 2/78 (2%)
Query 52 RSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGH 111
++QSR+E+K+RKA+L+LGMKP+ D+ +V I++AK FVI++PDVYKS + + YVIFG
Sbjct 58 NAKQSRSEKKSRKAVLKLGMKPVSDVSRVTIKRAKNVLFVISKPDVYKSPNAETYVIFGE 117
Query 112 A--EGVATQAHSEAAQRF 127
A + +++Q ++AAQRF
Sbjct 118 AKVDDLSSQLQTQAAQRF 135
> At4g10480
Length=212
Score = 92.0 bits (227), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 44/98 (44%), Positives = 71/98 (72%), Gaps = 4/98 (4%)
Query 53 SRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGHA 112
S+QSR+E+K+RKA+L+LGMKP+ D+ +V I+++K FVI++PDV+KS +++ YVIFG A
Sbjct 62 SKQSRSEKKSRKAMLKLGMKPVTDVSRVTIKRSKNVLFVISKPDVFKSPNSETYVIFGEA 121
Query 113 --EGVATQAHSEAAQRFTQGAVS--FGGGEGAAAAAAA 146
+ +++Q ++AAQRF V+ +G+ AA A
Sbjct 122 KIDDMSSQLQAQAAQRFKMPDVASMIPNTDGSEAATVA 159
> At3g12390
Length=203
Score = 91.3 bits (225), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 44/96 (45%), Positives = 71/96 (73%), Gaps = 4/96 (4%)
Query 52 RSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGH 111
+S+QSR+E+K+RKA+L+LGMKP+ + +V ++K+K FVI++PDV+KS ++D YVIFG
Sbjct 56 KSKQSRSEKKSRKAMLKLGMKPITGVSRVTVKKSKNILFVISKPDVFKSPASDTYVIFGE 115
Query 112 A--EGVATQAHSEAAQRFTQGAVS--FGGGEGAAAA 143
A E +++Q S+AA++F +S GE ++AA
Sbjct 116 AKIEDLSSQIQSQAAEQFKAPDLSNVISKGESSSAA 151
> At5g13850
Length=159
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 40/78 (51%), Positives = 63/78 (80%), Gaps = 2/78 (2%)
Query 52 RSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGH 111
RS+QSR+E+K+RKA+L+LGMKP+ + +V ++K+K FVI++PDV+KS ++D YVIFG
Sbjct 52 RSKQSRSEKKSRKAMLKLGMKPITGVSRVTVKKSKNILFVISKPDVFKSPASDTYVIFGE 111
Query 112 A--EGVATQAHSEAAQRF 127
A E +++Q S+AA++F
Sbjct 112 AKIEDLSSQLQSQAAEQF 129
> At3g49470
Length=217
Score = 83.6 bits (205), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 38/84 (45%), Positives = 61/84 (72%), Gaps = 2/84 (2%)
Query 46 GEGSPSRSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDA 105
+G S+QSR+E+K+RKA+L+LGMKP+ + +V I++ K F I++PDV+KS ++
Sbjct 60 AQGVSGSSKQSRSEKKSRKAMLKLGMKPVTGVSRVTIKRTKNVLFFISKPDVFKSPHSET 119
Query 106 YVIFGHA--EGVATQAHSEAAQRF 127
YVIFG A E +++Q ++AAQ+F
Sbjct 120 YVIFGEAKIEDLSSQLQTQAAQQF 143
> Hs5031931
Length=215
Score = 82.4 bits (202), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 37/79 (46%), Positives = 59/79 (74%), Gaps = 2/79 (2%)
Query 51 SRSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFG 110
S+++QSR+E+KARKA+ +LG++ + + +V IRK+K FVI +PDVYKS ++D Y++FG
Sbjct 65 SKAKQSRSEKKARKAMSKLGLRQVTGVTRVTIRKSKNILFVITKPDVYKSPASDTYIVFG 124
Query 111 HA--EGVATQAHSEAAQRF 127
A E ++ QA AA++F
Sbjct 125 EAKIEDLSQQAQLAAAEKF 143
> Hs20541584
Length=1469
Score = 82.0 bits (201), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 35/79 (44%), Positives = 60/79 (75%), Gaps = 2/79 (2%)
Query 51 SRSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFG 110
++++QSR+E+KARKA+ +LG++ + + ++ I+K+K FVIA+PDV+KS ++D YV+FG
Sbjct 1313 AKAKQSRSEKKARKAMSKLGLRQIQGVTRITIQKSKNILFVIAKPDVFKSPASDTYVVFG 1372
Query 111 HA--EGVATQAHSEAAQRF 127
A E ++ Q H AA++F
Sbjct 1373 EAKIEDLSQQVHKAAAEKF 1391
> 7303400
Length=217
Score = 81.3 bits (199), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 37/79 (46%), Positives = 56/79 (70%), Gaps = 2/79 (2%)
Query 51 SRSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFG 110
S+++QSR E+KARK +L+LG+K + + +V IRK+K FVI PDVYK+ +D Y++FG
Sbjct 65 SKAKQSRGEKKARKIMLKLGLKQIQGVNRVTIRKSKNILFVINNPDVYKNPHSDTYIVFG 124
Query 111 HA--EGVATQAHSEAAQRF 127
A E ++ QA AA++F
Sbjct 125 EAKIEDLSQQAQVAAAEKF 143
> Hs14042953
Length=213
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/79 (44%), Positives = 57/79 (72%), Gaps = 2/79 (2%)
Query 51 SRSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFG 110
S+++Q R+E+KARKA +LG++ + + +V IRK+K FVI +PDVYKS ++D Y++FG
Sbjct 64 SKAKQRRSEKKARKARFKLGLQQVTGVTRVTIRKSKNILFVITKPDVYKSPASDTYMVFG 123
Query 111 HA--EGVATQAHSEAAQRF 127
A E ++ +A AA++F
Sbjct 124 EAKIEDLSQEAQLAAAEKF 142
> CE22740
Length=195
Score = 71.6 bits (174), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 42/117 (35%), Positives = 64/117 (54%), Gaps = 19/117 (16%)
Query 7 AAEMANKKAEEPEHISSSSSDSGSEDEQKGEGAQQEQKE-----------GEGSPSRSRQ 55
+ E K+ +EP+ S SD+ E +QE E G+ +++Q
Sbjct 4 STETRQKEVKEPQVDVSDDSDN--------EAVEQELTEEQRRVAEAAGLGDHIDKQAKQ 55
Query 56 SRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGHA 112
SR+E+KARK +LG+K + + +V IRK+K FVI +PDV+KS +D Y+IFG A
Sbjct 56 SRSEKKARKLFSKLGLKQVTGVSRVCIRKSKNILFVINKPDVFKSPGSDTYIIFGEA 112
> 7296134
Length=341
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 32/82 (39%), Positives = 50/82 (60%), Gaps = 7/82 (8%)
Query 53 SRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGH- 111
S+QSR ERKAR+ +++L +KPM ++ +V ++K K I +PDVY+ + Y+ FG
Sbjct 180 SKQSRGERKARRILMKLDLKPMENVARVTMKKGKNILLYIDQPDVYRVAHSSTYIFFGEI 239
Query 112 ------AEGVATQAHSEAAQRF 127
VA+QA +AA+RF
Sbjct 240 CVEDTSTTSVASQAAVKAAERF 261
> Hs18588424
Length=215
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 33/78 (42%), Positives = 56/78 (71%), Gaps = 2/78 (2%)
Query 52 RSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGH 111
+++QSR+E++ARKA+ +LG+ + + +V I K+K FVI + DVYKS ++DAY++FG
Sbjct 66 KAKQSRSEKRARKAMSKLGLLQVTGVTRVTIWKSKNILFVITKLDVYKSPASDAYIVFGE 125
Query 112 A--EGVATQAHSEAAQRF 127
A + ++ QA AA++F
Sbjct 126 AKIQDLSQQAQLAAAEKF 143
> Hs18557511
Length=215
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 31/79 (39%), Positives = 54/79 (68%), Gaps = 2/79 (2%)
Query 51 SRSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFG 110
S+++Q +E+KA+KA+ +LG++ + + +V IRK+K FVI +P+V KS + D Y++ G
Sbjct 65 SKAKQIWSEKKAQKAMSKLGLRQVTGVTRVAIRKSKNILFVITKPNVCKSPALDTYIVSG 124
Query 111 HA--EGVATQAHSEAAQRF 127
A E ++ QA AA++F
Sbjct 125 EAKIEDLSEQAQLAAAEKF 143
> Hs17472280
Length=545
Score = 61.2 bits (147), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 30/79 (37%), Positives = 53/79 (67%), Gaps = 2/79 (2%)
Query 51 SRSRQSRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFG 110
S+++QS++E+KA+KA+ +LG++ + + +V I K+K FVI + DVYK + D Y++ G
Sbjct 403 SKAKQSQSEKKAQKAMSKLGLQQVTGVTRVTIWKSKNILFVITKADVYKRPALDTYIVLG 462
Query 111 --HAEGVATQAHSEAAQRF 127
E ++ QA AA++F
Sbjct 463 GTKNEDLSQQAQLAAAEKF 481
> YHR193c
Length=174
Score = 52.4 bits (124), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 23/58 (39%), Positives = 40/58 (68%), Gaps = 1/58 (1%)
Query 56 SRNERKARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGHAE 113
++NE+KAR+ I +LG+K +P I++V RK + I +P+V++S + YV+FG A+
Sbjct 14 NKNEKKARELIGKLGLKQIPGIIRVTFRKKDNQIYAIEKPEVFRSAGGN-YVVFGEAK 70
> SPBC25H2.05
Length=173
Score = 48.9 bits (115), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 26/63 (41%), Positives = 39/63 (61%), Gaps = 3/63 (4%)
Query 61 KARKAILRLGMKPMPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGH--AEGVATQ 118
KA+K I +LG+K + I +V +R+ K +I EP VYKS S +AY++ G E +A Q
Sbjct 26 KAQKLIQKLGLKRVEGITRVAMRRPKNILLIINEPIVYKS-SNNAYIVLGKVTVEDMAAQ 84
Query 119 AHS 121
A +
Sbjct 85 ARA 87
> Hs17463799
Length=282
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 77 IVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVI 108
+ +V I K+K FVI +PDVYKS +++ Y++
Sbjct 242 VTRVAIPKSKNILFVITKPDVYKSLASNTYIV 273
> At5g07530
Length=543
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 36/132 (27%), Positives = 56/132 (42%), Gaps = 10/132 (7%)
Query 10 MANKKAEEPEHISSSSSDSGSEDEQKGEGAQQEQKEGEGSPSRSRQSRNERKARKAILRL 69
M+ + +++P SS+ S S E + Q+EG PS S R+ RK +
Sbjct 1 MSEELSQKP---SSAQSLSLREGRNRFPFLSLSQREGRFFPSLSLSERDGRKFSFLSMFS 57
Query 70 GMKPMPDIVKVVIRKAKQSWFV------IAEPDVYKSTSTDAYVIFGHAEGVATQAHSEA 123
+ P+ +++K++I FV +A ST ++IF AT A
Sbjct 58 FLMPLLEVIKIIIASVASVIFVGFACVTLAGSAAALVVSTPVFIIFSPVLVPATIATVVL 117
Query 124 AQRFTQGAVSFG 135
A FT G SFG
Sbjct 118 ATGFTAGG-SFG 128
> Hs18559414_1
Length=377
Score = 33.5 bits (75), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 34/75 (45%), Gaps = 1/75 (1%)
Query 77 IVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGHAEG-VATQAHSEAAQRFTQGAVSFG 135
I+++ +K K + FVI PDVYKS ++D +G + +S R ++G
Sbjct 127 ILRLPSKKPKNTLFVIPRPDVYKSPASDTSTFWGKPRAKIHLSKYSYQMLRNSKGRQIQD 186
Query 136 GGEGAAAAAAAAAAR 150
G + A A R
Sbjct 187 GSRLSPLAPMPVALR 201
> Hs22044463
Length=195
Score = 33.5 bits (75), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 12/23 (52%), Positives = 18/23 (78%), Gaps = 0/23 (0%)
Query 84 KAKQSWFVIAEPDVYKSTSTDAY 106
K+K +FVI +PDVYK+ ++D Y
Sbjct 71 KSKNIFFVITKPDVYKNPASDTY 93
> Hs4505973
Length=289
Score = 32.0 bits (71), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 38/93 (40%), Gaps = 14/93 (15%)
Query 74 MPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGHAEGVATQAHSEAAQRFTQGAVS 133
+ DI K I + + + + +PD K +D Y+ + G A+R +G +S
Sbjct 120 LTDIAKYSIGRLRPHFLDVCDPDWSKINCSDGYIEYYICRG--------NAERVKEGRLS 171
Query 134 FGGGEGAAA------AAAAAAARTRGPWGPPLR 160
F G + + A AR +G W LR
Sbjct 172 FYSGHSSFSMYCMLFVALYLQARMKGDWARLLR 204
> Hs14721243
Length=285
Score = 31.2 bits (69), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 38/93 (40%), Gaps = 14/93 (15%)
Query 74 MPDIVKVVIRKAKQSWFVIAEPDVYKSTSTDAYVIFGHAEGVATQAHSEAAQRFTQGAVS 133
+ DI K I + + + + +PD K +D Y+ + G A+R +G +S
Sbjct 116 LTDIAKYSIGRLRPHFLDVCDPDWSKINCSDGYIEYYICRG--------NAERVKEGRLS 167
Query 134 FGGGEGAAA------AAAAAAARTRGPWGPPLR 160
F G + + A AR +G W LR
Sbjct 168 FYSGHSSFSMYCMLFVALYLQARMKGDWARLLR 200
> Hs18559237
Length=365
Score = 30.0 bits (66), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 18/66 (27%), Positives = 30/66 (45%), Gaps = 10/66 (15%)
Query 15 AEEPEHISSSSSDSGSEDEQKGEGAQQEQKEGEGSPSRSRQSRNERKARKAILRLGMKPM 74
A+ EH+ + SD + + E ++E K G G P K+ + LR G K
Sbjct 7 AKLEEHLENQPSDPTNTYARPAEPVEEENKNGNGKP----------KSLSSGLRKGTKKY 56
Query 75 PDIVKV 80
PD +++
Sbjct 57 PDYIQI 62
> YNL271c
Length=1953
Score = 30.0 bits (66), Expect = 2.6, Method: Composition-based stats.
Identities = 18/60 (30%), Positives = 30/60 (50%), Gaps = 1/60 (1%)
Query 8 AEMANKKAEEPEHISSSSSDSGSEDEQKGEGAQQEQKEGEGSPSRSRQSRNERKARKAIL 67
A+ N AEE E + E++QK +++QKE SPS +E + R+A++
Sbjct 1743 AQAQNLAAEEEERLYIKHKKI-VEEQQKRAQEKEKQKENSNSPSSEGNEEDEAEDRRAVM 1801
> Hs14727195
Length=709
Score = 29.6 bits (65), Expect = 3.0, Method: Composition-based stats.
Identities = 18/70 (25%), Positives = 35/70 (50%), Gaps = 10/70 (14%)
Query 19 EHISSSSSDSGSEDEQKGEGAQQEQK--------EGEGSPSRSRQSRNERK--ARKAILR 68
EH +S+ ++S + E K + ++++ EGEG+P + N+++ R A+
Sbjct 225 EHAASALTNSSASCESKNKNDSEKEQISSGHNMVEGEGAPKKKEPQENQKQQDVRTAMET 284
Query 69 LGMKPMPDIV 78
G+ P D V
Sbjct 285 TGLAPWQDGV 294
> Hs7706493
Length=709
Score = 29.6 bits (65), Expect = 3.1, Method: Composition-based stats.
Identities = 18/70 (25%), Positives = 35/70 (50%), Gaps = 10/70 (14%)
Query 19 EHISSSSSDSGSEDEQKGEGAQQEQK--------EGEGSPSRSRQSRNERK--ARKAILR 68
EH +S+ ++S + E K + ++++ EGEG+P + N+++ R A+
Sbjct 225 EHAASALTNSSASCESKNKNDSEKEQISSGHNMVEGEGAPKKKEPQENQKQQDVRTAMET 284
Query 69 LGMKPMPDIV 78
G+ P D V
Sbjct 285 TGLAPWQDGV 294
> HsM10047082
Length=848
Score = 28.1 bits (61), Expect = 8.4, Method: Composition-based stats.
Identities = 16/65 (24%), Positives = 31/65 (47%), Gaps = 5/65 (7%)
Query 2 GFFVLAAEMANKKAEEPEHISS-----SSSDSGSEDEQKGEGAQQEQKEGEGSPSRSRQS 56
G +L + + K++ P+ S S +D + E EG +QE+K GE ++
Sbjct 584 GIDLLKKDKSRKRSYSPDGKESPSDKKSKTDGSQKTESSTEGKEQEEKSGEDGEKDTKDD 643
Query 57 RNERK 61
+ E++
Sbjct 644 QTEQE 648
Lambda K H
0.310 0.123 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2598880752
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40