bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2872_orf1
Length=206
Score E
Sequences producing significant alignments: (Bits) Value
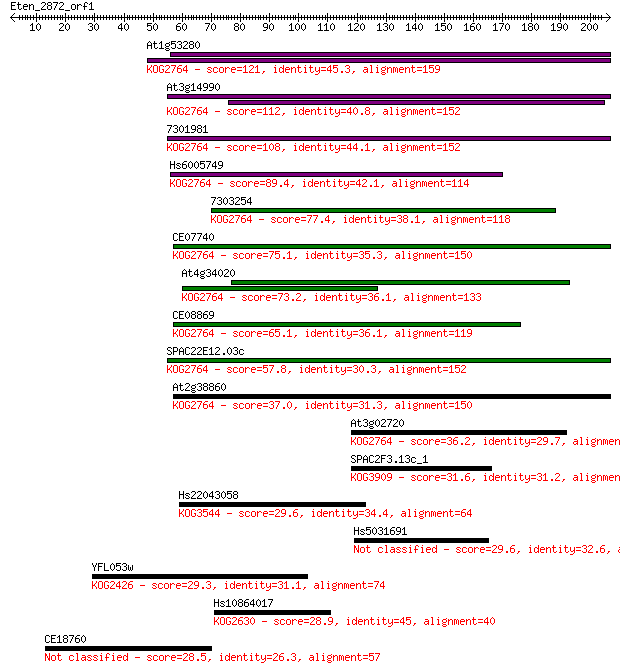
At1g53280 121 8e-28
At3g14990 112 4e-25
7301981 108 9e-24
Hs6005749 89.4 5e-18
7303254 77.4 2e-14
CE07740 75.1 9e-14
At4g34020 73.2 4e-13
CE08869 65.1 1e-10
SPAC22E12.03c 57.8 2e-08
At2g38860 37.0 0.028
At3g02720 36.2 0.049
SPAC2F3.13c_1 31.6 1.2
Hs22043058 29.6 4.0
Hs5031691 29.6 4.4
YFL053w 29.3 6.2
Hs10864017 28.9 7.3
CE18760 28.5 8.8
> At1g53280
Length=438
Score = 121 bits (304), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 70/153 (45%), Positives = 100/153 (65%), Gaps = 3/153 (1%)
Query 56 TKTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVS 115
T LV +A+G+E++E VA +DVL+RA V+VA++G+SL V S +K+ D+LL E +
Sbjct 257 TPQILVPIADGSEEMEAVAIIDVLKRAKANVVVAALGNSLEVVASRKVKLVADVLLDE-A 315
Query 116 EQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLENV 175
E+N+YD+IV+PGGL GAE S L +L+ Q S+K AICASPALV + G L+
Sbjct 316 EKNSYDLIVLPGGLGGAEAFASSEKLVNMLKKQAESNKPYGAICASPALVFEPHGLLKGK 375
Query 176 RAVAYPSFQQQLPLKG--EGRVCVDKHFITSVG 206
+A A+P+ +L + E RV VD + ITS G
Sbjct 376 KATAFPAMCSKLTDQSHIEHRVLVDGNLITSRG 408
Score = 109 bits (272), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 60/162 (37%), Positives = 92/162 (56%), Gaps = 4/162 (2%)
Query 48 LSASPPKMTKTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEG 107
+SA+ TK L+ +A+G E E V +DVLRR G V VASV + + V HG+K+
Sbjct 44 ISATMSSSTKKVLIPVAHGTEPFEAVVMIDVLRRGGADVTVASVENQVGVDACHGIKMVA 103
Query 108 DILLKEVSEQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQ 167
D LL ++++ + +D+I++PGGL G E ++ L +++ Q + + AAIC +PAL
Sbjct 104 DTLLSDITD-SVFDLIMLPGGLPGGETLKNCKPLEKMVKKQDTDGRLNAAICCAPALAFG 162
Query 168 QQGFLENVRAVAYPSFQQQLPLKG---EGRVCVDKHFITSVG 206
G LE +A YP F ++L E RV +D +TS G
Sbjct 163 TWGLLEGKKATCYPVFMEKLAACATAVESRVEIDGKIVTSRG 204
> At3g14990
Length=392
Score = 112 bits (281), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 62/156 (39%), Positives = 90/156 (57%), Gaps = 5/156 (3%)
Query 55 MTKTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEV 114
TKT L+ +A+G E +E VA + VLRR G V VASV + V HG+K+ D LL ++
Sbjct 4 FTKTVLIPIAHGTEPLEAVAMITVLRRGGADVTVASVETQVGVDACHGIKMVADTLLSDI 63
Query 115 SEQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLEN 174
++ + +D+IV+PGGL G E ++ L +++ Q S + AAIC +PAL L G LE
Sbjct 64 TD-SVFDLIVLPGGLPGGETLKNCKSLENMVKKQDSDGRLNAAICCAPALALGTWGLLEG 122
Query 175 VRAVAYPSFQQQLPLKG----EGRVCVDKHFITSVG 206
+A YP F ++L E RV +D +TS G
Sbjct 123 KKATGYPVFMEKLAATCATAVESRVQIDGRIVTSRG 158
Score = 97.1 bits (240), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 55/131 (41%), Positives = 80/131 (61%), Gaps = 3/131 (2%)
Query 76 VDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVSEQNNYDVIVIPGGLKGAENC 135
VD+LRRA V++A+VG+SL V+ S K+ ++LL EV+E++ +D+IV+PGGL GA+
Sbjct 231 VDILRRAKANVVIAAVGNSLEVEGSRKAKLVAEVLLDEVAEKS-FDLIVLPGGLNGAQRF 289
Query 136 RDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLENVRAVAYPSFQQQLPLKG--EG 193
L +LR Q ++K ICASPA V + G L+ +A +P +L K E
Sbjct 290 ASCEKLVNMLRKQAEANKPYGGICASPAYVFEPNGLLKGKKATTHPVVSDKLSDKSHIEH 349
Query 194 RVCVDKHFITS 204
RV VD + ITS
Sbjct 350 RVVVDGNVITS 360
> 7301981
Length=187
Score = 108 bits (269), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 67/156 (42%), Positives = 93/156 (59%), Gaps = 6/156 (3%)
Query 55 MTKTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEV 114
M+K+ALV+LA GAE++EF+ A DVLRRAG+KV VA + VK S ++I D L +V
Sbjct 1 MSKSALVILAPGAEEMEFIIAADVLRRAGIKVTVAGLNGGEAVKCSRDVQILPDTSLAQV 60
Query 115 SEQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLEN 174
+ + +DV+V+PGGL G+ +SS + LLR+Q S IAAICA+P VL + G
Sbjct 61 A-SDKFDVVVLPGGLGGSNAMGESSLVGDLLRSQESGGGLIAAICAAPT-VLAKHGVASG 118
Query 175 VRAVAYPSFQQQL----PLKGEGRVCVDKHFITSVG 206
+YPS + QL + V D + ITS G
Sbjct 119 KSLTSYPSMKPQLVNNYSYVDDKTVVKDGNLITSRG 154
> Hs6005749
Length=189
Score = 89.4 bits (220), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 48/114 (42%), Positives = 72/114 (63%), Gaps = 0/114 (0%)
Query 56 TKTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVS 115
+K ALV+LA GAE++E V VDV+RRAG+KV VA + V+ S + I D L++
Sbjct 3 SKRALVILAKGAEEMETVIPVDVMRRAGIKVTVAGLAGKDPVQCSRDVVICPDASLEDAK 62
Query 116 EQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQ 169
++ YDV+V+PGG GA+N +S+ + +L+ Q + IAAICA P +L +
Sbjct 63 KEGPYDVVVLPGGNLGAQNLSESAAVKEILKEQENRKGLIAAICAGPTALLAHE 116
> 7303254
Length=174
Score = 77.4 bits (189), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 45/118 (38%), Positives = 65/118 (55%), Gaps = 1/118 (0%)
Query 70 VEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVSEQNNYDVIVIPGGL 129
+EF + DVLRR + V VA + D VK S + I D L+E + +YDV+V+PGGL
Sbjct 1 MEFTISADVLRRGKILVTVAGLHDCEPVKCSRSVVIVPDTSLEEAVTRGDYDVVVLPGGL 60
Query 130 KGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLENVRAVAYPSFQQQL 187
G + +SS + +LR Q S IAAICA+P L + G + ++P + QL
Sbjct 61 AGNKALMNSSAVGDVLRCQESKGGLIAAICAAPT-ALAKHGIGKGKSITSHPDMKPQL 117
> CE07740
Length=187
Score = 75.1 bits (183), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 53/155 (34%), Positives = 86/155 (55%), Gaps = 8/155 (5%)
Query 57 KTALVVLA-NGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVS 115
K+AL++LA GAE++E + DVL R ++V+ A + + VK + G I D+ L++V
Sbjct 4 KSALIILAAEGAEEMEVIITGDVLARGEIRVVYAGLDGAEPVKCARGAHIVPDVKLEDV- 62
Query 116 EQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLENV 175
E +D++++PGG G+ +S + +L++Q S I AICA+P +L E V
Sbjct 63 ETEKFDIVILPGGQPGSNTLAESLLVRDVLKSQVESGGLIGAICAAPIALLSHGVKAELV 122
Query 176 RAVAYPSFQQQLPLKG----EGRVCVDKHFITSVG 206
++PS +++L G E RV V ITS G
Sbjct 123 --TSHPSVKEKLEKGGYKYSEDRVVVSGKIITSRG 155
> At4g34020
Length=334
Score = 73.2 bits (178), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 40/116 (34%), Positives = 63/116 (54%), Gaps = 1/116 (0%)
Query 77 DVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVSEQNNYDVIVIPGGLKGAENCR 136
DVLRRAG V VASV L V+ S G ++ D+L+ + ++Q YD++ +PGG+ GA R
Sbjct 116 DVLRRAGADVTVASVEQKLEVEGSSGTRLLADVLISKCADQV-YDLVALPGGMPGAVRLR 174
Query 137 DSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLENVRAVAYPSFQQQLPLKGE 192
D L +++ Q + AI +PA+ L G L R + + + + + GE
Sbjct 175 DCEILEKIMKRQAEDKRLYGAISMAPAITLLPWGLLTRKRLPTFWAVKTNIQISGE 230
Score = 55.5 bits (132), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 31/67 (46%), Positives = 45/67 (67%), Gaps = 1/67 (1%)
Query 60 LVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVSEQNN 119
L+ +ANG+E VE V+ DVLRRA + V V+SV SL + G KI D L+ E +E ++
Sbjct 266 LIPVANGSEAVELVSIADVLRRAKVDVTVSSVERSLRITAFQGTKIITDKLIGEAAE-SS 324
Query 120 YDVIVIP 126
YD+I++P
Sbjct 325 YDLIILP 331
> CE08869
Length=186
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 43/120 (35%), Positives = 65/120 (54%), Gaps = 4/120 (3%)
Query 57 KTALVVLA-NGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDILLKEVS 115
K+AL++L AE++E + DVL R G++VL A + VK + G +I D+ LK+V
Sbjct 5 KSALILLPPEDAEEIEVIVTGDVLVRGGLQVLYAG-SSTEPVKCAKGARIVPDVALKDV- 62
Query 116 EQNNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLENV 175
+ +D+I+IPGG G + + LL+ Q S I AICA P ++L E V
Sbjct 63 KNKTFDIIIIPGG-PGCSKLAECPVIGELLKTQVKSGGLIGAICAGPTVLLAHGIVAERV 121
> SPAC22E12.03c
Length=191
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 46/163 (28%), Positives = 82/163 (50%), Gaps = 14/163 (8%)
Query 55 MTKTALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDS--LFVKTSHGLKIEGDILLK 112
M K L V A+G +++EF A + +RA + + VG++ VK S +++ + K
Sbjct 1 MVKVCLFV-ADGTDEIEFSAPWGIFKRAEIPIDSVYVGENKDRLVKMSRDVEMYANRSYK 59
Query 113 EVSEQNN----YDVIVIPGGLKGAENCRDSSHLAALLRAQHSS-SKWIAAICASPALVLQ 167
E+ ++ YD+ +IPGG GA+ + + +++ + +KWI ICA L +
Sbjct 60 EIPSADDFAKQYDIAIIPGGGLGAKTLSTTPFVQQVVKEFYKKPNKWIGMICAG-TLTAK 118
Query 168 QQGFLENVRAVAYPSFQQQLPLKG----EGRVCVDKHFITSVG 206
G L N + +PS + QL G + V ++++ ITS G
Sbjct 119 TSG-LPNKQITGHPSVRGQLEEGGYKYLDQPVVLEENLITSQG 160
> At2g38860
Length=398
Score = 37.0 bits (84), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 47/184 (25%), Positives = 79/184 (42%), Gaps = 41/184 (22%)
Query 57 KTALVVLANGAEDVEFVAAVDVLRRAGMKVLVAS----VGDSLFVKTSH---GLKIEGDI 109
K+AL++ + E E + + VL+ G+ V S GD V ++H GL++ ++
Sbjct 7 KSALLLCGDYMEAYETIVPLYVLQSFGVSVHCVSPNRNAGDRC-VMSAHDFLGLELYTEL 65
Query 110 LLKEVS--------EQNNYDVIVIPGG-----LKGAENCRDSSHLAALLRAQHSSSKWIA 156
++ +++ NYDVI+IPGG L E C D L+ S K I
Sbjct 66 VVDQLTLNANFDDVTPENYDVIIIPGGRFTELLSADEKCVD------LVARFAESKKLIF 119
Query 157 AICASPALVLQQQGFLENVRAVAYPSFQQQLPLKG-------------EGRVCV-DKHFI 202
C S +++ V+ A+ S + + L G E CV D +F+
Sbjct 120 TSCHSQVMLMAAGILAGGVKCTAFESIKPLIELSGGEWWQQPGIQSMFEITDCVKDGNFM 179
Query 203 TSVG 206
++VG
Sbjct 180 STVG 183
> At3g02720
Length=388
Score = 36.2 bits (82), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 40/74 (54%), Gaps = 2/74 (2%)
Query 118 NNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALVLQQQGFLENVRA 177
++YD +VIPGG + E + H+ +++ +S K +A+IC +L G L+ +
Sbjct 273 SSYDALVIPGG-RAPEYLALNEHVLNIVKEFMNSEKPVASICHGQQ-ILAAAGVLKGRKC 330
Query 178 VAYPSFQQQLPLKG 191
AYP+ + + L G
Sbjct 331 TAYPAVKLNVVLGG 344
> SPAC2F3.13c_1
Length=484
Score = 31.6 bits (70), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 23/48 (47%), Gaps = 0/48 (0%)
Query 118 NNYDVIVIPGGLKGAENCRDSSHLAALLRAQHSSSKWIAAICASPALV 165
+N + IP G E+C D + +L + + KWIAA P L+
Sbjct 44 DNVEEFDIPALYVGLEDCLDRLEASPILTNEGTIKKWIAAPSVQPTLL 91
> Hs22043058
Length=3631
Score = 29.6 bits (65), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 32/68 (47%), Gaps = 7/68 (10%)
Query 59 ALVVLANGAEDVEFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGD----ILLKEV 114
LVV+ +G + AV L+ G+ VLV +GD V H L I G+ I ++
Sbjct 2030 TLVVITSGDPRYDVADAVKTLKDLGICVLVLGIGD---VYKEHLLPITGNSEKIITFQDF 2086
Query 115 SEQNNYDV 122
+ N DV
Sbjct 2087 DKLKNVDV 2094
> Hs5031691
Length=268
Score = 29.6 bits (65), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 15/56 (26%), Positives = 25/56 (44%), Gaps = 10/56 (17%)
Query 119 NYDVIVIPGGLKGAEN----------CRDSSHLAALLRAQHSSSKWIAAICASPAL 164
N+D + PGG A+N C+ + + +L+ H + K I C +P L
Sbjct 127 NHDAAIFPGGFGAAKNLSTFAVDGKDCKVNKEVERVLKEFHQAGKPIGLCCIAPVL 182
> YFL053w
Length=591
Score = 29.3 bits (64), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 38/76 (50%), Gaps = 5/76 (6%)
Query 29 SLSKAASFATQQQQQQQQQLSA-SPPKMTKTALVVLANGAEDV-EFVAAVDVLRRAGMKV 86
+++ A FA+ +Q + A PK T L+++ N D+ F A + + AGMKV
Sbjct 76 AIAAGAIFASPSTKQIYSAIKAVESPKGT---LIIVKNYTGDIIHFGLAAERAKAAGMKV 132
Query 87 LVASVGDSLFVKTSHG 102
+ +VGD + V G
Sbjct 133 ELVAVGDDVSVGKKKG 148
> Hs10864017
Length=261
Score = 28.9 bits (63), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 20/40 (50%), Gaps = 0/40 (0%)
Query 71 EFVAAVDVLRRAGMKVLVASVGDSLFVKTSHGLKIEGDIL 110
+ V AV R AGMKV + S G K G EGDIL
Sbjct 134 DVVPAVRKWREAGMKVYIYSSGSVEAQKLLFGHSTEGDIL 173
> CE18760
Length=715
Score = 28.5 bits (62), Expect = 8.8, Method: Composition-based stats.
Identities = 15/57 (26%), Positives = 27/57 (47%), Gaps = 0/57 (0%)
Query 13 LNPIHFNNSNLASFPASLSKAASFATQQQQQQQQQLSASPPKMTKTALVVLANGAED 69
L+P FN + F AS+SK F + ++ Q +A+ P + +L+ + D
Sbjct 467 LHPTRFNTESTQLFVASISKLLDFRNESERIMLFQATANTPTLMDLISSILSRPSTD 523
Lambda K H
0.316 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3694581778
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40