bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
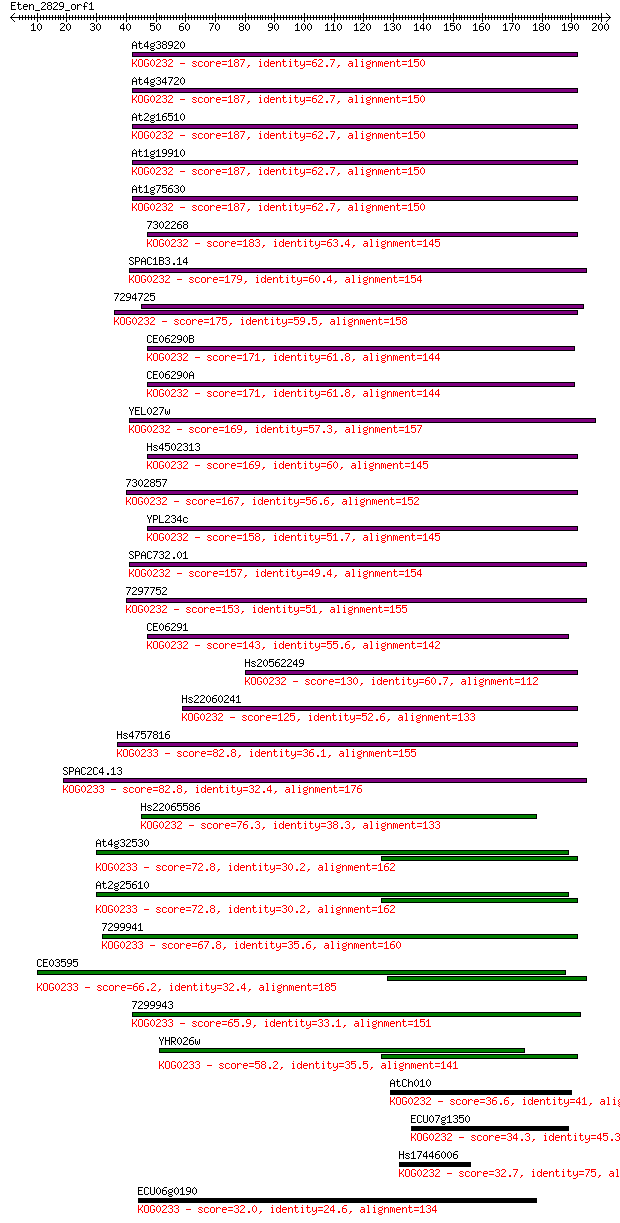
Query= Eten_2829_orf1
Length=202
Score E
Sequences producing significant alignments: (Bits) Value
At4g38920 187 1e-47
At4g34720 187 1e-47
At2g16510 187 1e-47
At1g19910 187 1e-47
At1g75630 187 1e-47
7302268 183 2e-46
SPAC1B3.14 179 4e-45
7294725 175 5e-44
CE06290B 171 1e-42
CE06290A 171 1e-42
YEL027w 169 3e-42
Hs4502313 169 5e-42
7302857 167 2e-41
YPL234c 158 6e-39
SPAC732.01 157 2e-38
7297752 153 3e-37
CE06291 143 2e-34
Hs20562249 130 2e-30
Hs22060241 125 5e-29
Hs4757816 82.8 4e-16
SPAC2C4.13 82.8 4e-16
Hs22065586 76.3 4e-14
At4g32530 72.8 4e-13
At2g25610 72.8 4e-13
7299941 67.8 1e-11
CE03595 66.2 4e-11
7299943 65.9 5e-11
YHR026w 58.2 1e-08
AtCh010 36.6 0.035
ECU07g1350 34.3 0.17
Hs17446006 32.7 0.49
ECU06g0190 32.0 0.93
> At4g38920
Length=164
Score = 187 bits (475), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 94/151 (62%), Positives = 126/151 (83%), Gaps = 1/151 (0%)
Query 42 DPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGI 101
D + FFGF+G +A+VF+ +GAAYGTAKSGVGV+S+GVMRP+L+M+SI+PV+MAG+LGI
Sbjct 7 DETAPFFGFLGAAAALVFSCMGAAYGTAKSGVGVASMGVMRPELVMKSIVPVVMAGVLGI 66
Query 102 YGLIISIVISS-VMGSPDVYSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQ 160
YGLII+++IS+ + Y + G+ HL+ GLA GL+ L+AG+AIGIVGDAGVRANAQQ
Sbjct 67 YGLIIAVIISTGINPKAKSYYLFDGYAHLSSGLACGLAGLSAGMAIGIVGDAGVRANAQQ 126
Query 161 PKLFFGVTLILIFAEAIGLYGLIIALVIVTR 191
PKLF G+ LILIFAEA+ LYGLI+ +++ +R
Sbjct 127 PKLFVGMILILIFAEALALYGLIVGIILSSR 157
> At4g34720
Length=164
Score = 187 bits (475), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 94/151 (62%), Positives = 126/151 (83%), Gaps = 1/151 (0%)
Query 42 DPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGI 101
D + FFGF+G +A+VF+ +GAAYGTAKSGVGV+S+GVMRP+L+M+SI+PV+MAG+LGI
Sbjct 7 DETAPFFGFLGAAAALVFSCMGAAYGTAKSGVGVASMGVMRPELVMKSIVPVVMAGVLGI 66
Query 102 YGLIISIVISS-VMGSPDVYSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQ 160
YGLII+++IS+ + Y + G+ HL+ GLA GL+ L+AG+AIGIVGDAGVRANAQQ
Sbjct 67 YGLIIAVIISTGINPKAKSYYLFDGYAHLSSGLACGLAGLSAGMAIGIVGDAGVRANAQQ 126
Query 161 PKLFFGVTLILIFAEAIGLYGLIIALVIVTR 191
PKLF G+ LILIFAEA+ LYGLI+ +++ +R
Sbjct 127 PKLFVGMILILIFAEALALYGLIVGIILSSR 157
> At2g16510
Length=164
Score = 187 bits (475), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 94/151 (62%), Positives = 126/151 (83%), Gaps = 1/151 (0%)
Query 42 DPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGI 101
D + FFGF+G +A+VF+ +GAAYGTAKSGVGV+S+GVMRP+L+M+SI+PV+MAG+LGI
Sbjct 7 DETAPFFGFLGAAAALVFSCMGAAYGTAKSGVGVASMGVMRPELVMKSIVPVVMAGVLGI 66
Query 102 YGLIISIVISS-VMGSPDVYSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQ 160
YGLII+++IS+ + Y + G+ HL+ GLA GL+ L+AG+AIGIVGDAGVRANAQQ
Sbjct 67 YGLIIAVIISTGINPKAKSYYLFDGYAHLSSGLACGLAGLSAGMAIGIVGDAGVRANAQQ 126
Query 161 PKLFFGVTLILIFAEAIGLYGLIIALVIVTR 191
PKLF G+ LILIFAEA+ LYGLI+ +++ +R
Sbjct 127 PKLFVGMILILIFAEALALYGLIVGIILSSR 157
> At1g19910
Length=165
Score = 187 bits (475), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 94/151 (62%), Positives = 126/151 (83%), Gaps = 1/151 (0%)
Query 42 DPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGI 101
D + FFGF+G +A+VF+ +GAAYGTAKSGVGV+S+GVMRP+L+M+SI+PV+MAG+LGI
Sbjct 8 DETAPFFGFLGAAAALVFSCMGAAYGTAKSGVGVASMGVMRPELVMKSIVPVVMAGVLGI 67
Query 102 YGLIISIVISS-VMGSPDVYSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQ 160
YGLII+++IS+ + Y + G+ HL+ GLA GL+ L+AG+AIGIVGDAGVRANAQQ
Sbjct 68 YGLIIAVIISTGINPKAKSYYLFDGYAHLSSGLACGLAGLSAGMAIGIVGDAGVRANAQQ 127
Query 161 PKLFFGVTLILIFAEAIGLYGLIIALVIVTR 191
PKLF G+ LILIFAEA+ LYGLI+ +++ +R
Sbjct 128 PKLFVGMILILIFAEALALYGLIVGIILSSR 158
> At1g75630
Length=166
Score = 187 bits (475), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 94/151 (62%), Positives = 126/151 (83%), Gaps = 1/151 (0%)
Query 42 DPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGI 101
D + FFGF+G +A+VF+ +GAAYGTAKSGVGV+S+GVMRP+L+M+SI+PV+MAG+LGI
Sbjct 9 DETAPFFGFLGAAAALVFSCMGAAYGTAKSGVGVASMGVMRPELVMKSIVPVVMAGVLGI 68
Query 102 YGLIISIVISS-VMGSPDVYSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQ 160
YGLII+++IS+ + Y + G+ HL+ GLA GL+ L+AG+AIGIVGDAGVRANAQQ
Sbjct 69 YGLIIAVIISTGINPKAKSYYLFDGYAHLSSGLACGLAGLSAGMAIGIVGDAGVRANAQQ 128
Query 161 PKLFFGVTLILIFAEAIGLYGLIIALVIVTR 191
PKLF G+ LILIFAEA+ LYGLI+ +++ +R
Sbjct 129 PKLFVGMILILIFAEALALYGLIVGIILSSR 159
> 7302268
Length=159
Score = 183 bits (464), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 92/145 (63%), Positives = 117/145 (80%), Gaps = 0/145 (0%)
Query 47 FFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLII 106
FFG MG SAI+F+ LGAAYGTAKSG G++++ VMRP+LIM+SI+PV+MAGI+ IYGL++
Sbjct 15 FFGVMGAASAIIFSALGAAYGTAKSGTGIAAMSVMRPELIMKSIIPVVMAGIIAIYGLVV 74
Query 107 SIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQPKLFFG 166
+++I+ + P YS Y GF HL GLAVG S LAAG AIGIVGDAGVR AQQP+LF G
Sbjct 75 AVLIAGALEEPSKYSLYRGFIHLGAGLAVGFSGLAAGFAIGIVGDAGVRGTAQQPRLFVG 134
Query 167 VTLILIFAEAIGLYGLIIALVIVTR 191
+ LILIFAE +GLYGLI+A+ + T+
Sbjct 135 MILILIFAEVLGLYGLIVAIYLYTK 159
> SPAC1B3.14
Length=161
Score = 179 bits (453), Expect = 4e-45, Method: Compositional matrix adjust.
Identities = 93/154 (60%), Positives = 122/154 (79%), Gaps = 2/154 (1%)
Query 41 CDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILG 100
C + FFG MG T+AIVFA+ GAAYGTAK+GVG+S++GV+RPDLI+++ +PV+MAGI+
Sbjct 6 CPVYAPFFGVMGCTAAIVFASFGAAYGTAKAGVGISAMGVLRPDLIVKNTIPVVMAGIIA 65
Query 101 IYGLIISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQ 160
IYGL++S++IS + + S Y+GF L GL+VGL+ LAAG AIGIVGDAGVR AQQ
Sbjct 66 IYGLVVSVLISGNL--KQILSLYSGFIQLGAGLSVGLAGLAAGFAIGIVGDAGVRGTAQQ 123
Query 161 PKLFFGVTLILIFAEAIGLYGLIIALVIVTRKVD 194
P+LF + LILIFAE +GLYGLI+AL++ TR D
Sbjct 124 PRLFVAMILILIFAEVLGLYGLIVALLLNTRATD 157
> 7294725
Length=317
Score = 175 bits (444), Expect = 5e-44, Method: Compositional matrix adjust.
Identities = 91/149 (61%), Positives = 119/149 (79%), Gaps = 2/149 (1%)
Query 45 SVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGL 104
S FFG MG SAI+F+ LGAAYGTAKSG G++++ VMRP+LIM+SI+PV+MAGI+ IYGL
Sbjct 14 SFFFGSMGAASAIIFSALGAAYGTAKSGTGIAAMAVMRPELIMKSIIPVVMAGIIAIYGL 73
Query 105 IISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQPKLF 164
++S++I+ + D Y+ G+ HLA GL+VG + LAAG AIGIVGDAGVR AQQP+LF
Sbjct 74 VVSVLIAGSL--SDSYTIRKGYIHLAAGLSVGFAGLAAGFAIGIVGDAGVRGTAQQPRLF 131
Query 165 FGVTLILIFAEAIGLYGLIIALVIVTRKV 193
G+ LILIFAE +GLYGLI+A+ + T K+
Sbjct 132 VGMILILIFAEVLGLYGLIVAIYLYTNKM 160
Score = 156 bits (395), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 83/157 (52%), Positives = 115/157 (73%), Gaps = 3/157 (1%)
Query 36 SAFLQCDPNSVFF-GFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVI 94
+A L +P+ FF G G AI+F LGA+YGTA SGVG++ + V RPD+IM++I+PV+
Sbjct 162 TAALNEEPSYAFFLGCTGAAVAIIFTTLGASYGTAVSGVGIAKMAVNRPDMIMKAIIPVV 221
Query 95 MAGILGIYGLIISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGV 154
MAGI+ IYGL++S++I+ +G D Y+ + HL GL+VGL L AG+AIGI GDAGV
Sbjct 222 MAGIIAIYGLVVSVLIAGSIG--DDYTMEDSYVHLGAGLSVGLPGLTAGVAIGIAGDAGV 279
Query 155 RANAQQPKLFFGVTLILIFAEAIGLYGLIIALVIVTR 191
R A+QP+LF G+ LILIFAE + LYGLI+A+ + T+
Sbjct 280 RGTAEQPRLFVGMVLILIFAEVLALYGLIVAIYLYTK 316
> CE06290B
Length=161
Score = 171 bits (432), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 89/145 (61%), Positives = 112/145 (77%), Gaps = 1/145 (0%)
Query 47 FFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLII 106
FFG+MG SA +F LGAAYGTAKS VG+ S+GVMRP+LIM+S++PVIMAGI+GIYGL++
Sbjct 16 FFGYMGAASAQIFTVLGAAYGTAKSAVGICSMGVMRPELIMKSVIPVIMAGIIGIYGLVV 75
Query 107 SIVISSVMGSPDV-YSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQPKLFF 165
++V+ + S Y GF HLA GL GL L AG AIGIVGDAGVR AQQP+LF
Sbjct 76 AMVLKGKVTSASAGYDLNKGFAHLAAGLTCGLCGLGAGYAIGIVGDAGVRGTAQQPRLFV 135
Query 166 GVTLILIFAEAIGLYGLIIALVIVT 190
G+ LILIF+E +GLYG+I+AL++ T
Sbjct 136 GMILILIFSEVLGLYGMIVALILGT 160
> CE06290A
Length=161
Score = 171 bits (432), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 89/145 (61%), Positives = 112/145 (77%), Gaps = 1/145 (0%)
Query 47 FFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLII 106
FFG+MG SA +F LGAAYGTAKS VG+ S+GVMRP+LIM+S++PVIMAGI+GIYGL++
Sbjct 16 FFGYMGAASAQIFTVLGAAYGTAKSAVGICSMGVMRPELIMKSVIPVIMAGIIGIYGLVV 75
Query 107 SIVISSVMGSPDV-YSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQPKLFF 165
++V+ + S Y GF HLA GL GL L AG AIGIVGDAGVR AQQP+LF
Sbjct 76 AMVLKGKVTSASAGYDLNKGFAHLAAGLTCGLCGLGAGYAIGIVGDAGVRGTAQQPRLFV 135
Query 166 GVTLILIFAEAIGLYGLIIALVIVT 190
G+ LILIF+E +GLYG+I+AL++ T
Sbjct 136 GMILILIFSEVLGLYGMIVALILGT 160
> YEL027w
Length=160
Score = 169 bits (429), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 90/158 (56%), Positives = 120/158 (75%), Gaps = 3/158 (1%)
Query 41 CDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILG 100
C + FFG +G SAI+F +LGAAYGTAKSGVG+ + V+RPDL+ ++I+PVIMAGI+
Sbjct 5 CPVYAPFFGAIGCASAIIFTSLGAAYGTAKSGVGICATCVLRPDLLFKNIVPVIMAGIIA 64
Query 101 IYGLIISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQ 160
IYGL++S+++ +G + Y GF L GL+VGLS LAAG AIGIVGDAGVR ++QQ
Sbjct 65 IYGLVVSVLVCYSLGQKQ--ALYTGFIQLGAGLSVGLSGLAAGFAIGIVGDAGVRGSSQQ 122
Query 161 PKLFFGVTLILIFAEAIGLYGLIIALVIVTRKV-DGLC 197
P+LF G+ LILIFAE +GLYGLI+AL++ +R D +C
Sbjct 123 PRLFVGMILILIFAEVLGLYGLIVALLLNSRATQDVVC 160
> Hs4502313
Length=155
Score = 169 bits (427), Expect = 5e-42, Method: Compositional matrix adjust.
Identities = 87/145 (60%), Positives = 116/145 (80%), Gaps = 2/145 (1%)
Query 47 FFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLII 106
FF MG ++A+VF+ LGAAYGTAKSG G++++ VMRP+ IM+SI+PV+MAGI+ IYGL++
Sbjct 13 FFAVMGASAAMVFSALGAAYGTAKSGTGIAAMSVMRPEQIMKSIIPVVMAGIIAIYGLVV 72
Query 107 SIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQPKLFFG 166
+++I++ + D S Y F L GL+VGLS LAAG AIGIVGDAGVR AQQP+LF G
Sbjct 73 AVLIANSLN--DDISLYKSFLQLGAGLSVGLSGLAAGFAIGIVGDAGVRGTAQQPRLFVG 130
Query 167 VTLILIFAEAIGLYGLIIALVIVTR 191
+ LILIFAE +GLYGLI+AL++ T+
Sbjct 131 MILILIFAEVLGLYGLIVALILSTK 155
> 7302857
Length=155
Score = 167 bits (422), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 86/152 (56%), Positives = 119/152 (78%), Gaps = 5/152 (3%)
Query 40 QCDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGIL 99
QC FF +G AIVF+ LGAAYGTAK+ VG+SS+ + P LIM++I+PV+MAGI+
Sbjct 9 QCAS---FFCILGAVCAIVFSTLGAAYGTAKASVGISSMSIKHPQLIMKAIVPVVMAGII 65
Query 100 GIYGLIISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQ 159
IYGL+I+++++ + SP YSAY GF +L+ GLAVG+S + AG+AIG+VG+AGVRA+AQ
Sbjct 66 AIYGLVIAVLLAGSLSSP--YSAYKGFLNLSAGLAVGVSGMGAGIAIGVVGEAGVRASAQ 123
Query 160 QPKLFFGVTLILIFAEAIGLYGLIIALVIVTR 191
QPKLF + LILIFAE +GLYGLI+A+ + ++
Sbjct 124 QPKLFVAIILILIFAEVLGLYGLIVAIYLFSK 155
> YPL234c
Length=164
Score = 158 bits (400), Expect = 6e-39, Method: Compositional matrix adjust.
Identities = 75/145 (51%), Positives = 112/145 (77%), Gaps = 0/145 (0%)
Query 47 FFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLII 106
FFGF G +A+V + LGAA GTAKSG+G++ +G +P+LIM+S++PV+M+GIL IYGL++
Sbjct 17 FFGFAGCAAAMVLSCLGAAIGTAKSGIGIAGIGTFKPELIMKSLIPVVMSGILAIYGLVV 76
Query 107 SIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQPKLFFG 166
+++I+ + + Y+ + GF HL+ GL VG + L++G AIG+VGD GVR QP+LF G
Sbjct 77 AVLIAGNLSPTEDYTLFNGFMHLSCGLCVGFACLSSGYAIGMVGDVGVRKYMHQPRLFVG 136
Query 167 VTLILIFAEAIGLYGLIIALVIVTR 191
+ LILIF+E +GLYG+I+AL++ TR
Sbjct 137 IVLILIFSEVLGLYGMIVALILNTR 161
> SPAC732.01
Length=162
Score = 157 bits (396), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 76/154 (49%), Positives = 119/154 (77%), Gaps = 0/154 (0%)
Query 41 CDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILG 100
C S FFGF G+ +++VF+ LGA YGTA +G G+++VG RP+++M+S++PV+M+GI+G
Sbjct 6 CPIYSSFFGFAGVCASMVFSCLGAGYGTALAGRGIAAVGAFRPEIVMKSLIPVVMSGIIG 65
Query 101 IYGLIISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQ 160
+YGL++S++I+ M + YS ++GF HL+ GLAVGL+ +AAG AIG+VGD GV++ +Q
Sbjct 66 VYGLVMSVLIAGDMSPDNDYSLFSGFIHLSAGLAVGLTGVAAGYAIGVVGDRGVQSFMRQ 125
Query 161 PKLFFGVTLILIFAEAIGLYGLIIALVIVTRKVD 194
++F + LILIFAE +GLYGLI+ L++ T+ +
Sbjct 126 DRIFVSMVLILIFAEVLGLYGLIVGLILQTKTSN 159
> 7297752
Length=193
Score = 153 bits (386), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 79/155 (50%), Positives = 110/155 (70%), Gaps = 0/155 (0%)
Query 40 QCDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGIL 99
+ P S F+G MG+ + V + GAAYGTA SG G+++ VMRP+L+M+SI+PV+MAGI+
Sbjct 39 RYPPYSPFYGVMGVVFSSVLTSAGAAYGTAVSGTGIAATAVMRPELVMKSIIPVVMAGII 98
Query 100 GIYGLIISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQ 159
IYGL++S+++S + YS G+ HLA GL+VG + LAAG A+G VG+ GVR A
Sbjct 99 AIYGLVVSVLLSGELAPAPKYSLPTGYVHLAAGLSVGFAGLAAGYAVGEVGEVGVRHIAL 158
Query 160 QPKLFFGVTLILIFAEAIGLYGLIIALVIVTRKVD 194
QP+LF G+ LILIFAE +GLYGLII + + T +
Sbjct 159 QPRLFIGMILILIFAEVLGLYGLIIGIYLYTVNIK 193
> CE06291
Length=169
Score = 143 bits (361), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 79/143 (55%), Positives = 110/143 (76%), Gaps = 1/143 (0%)
Query 47 FFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLII 106
FFG +G+TSA+ FA G+AYGTAK+G G++S+ V RPDL+M++I+PV+MAGI+ IYGL++
Sbjct 24 FFGSLGVTSAMAFAAAGSAYGTAKAGTGIASMAVARPDLVMKAIIPVVMAGIVAIYGLVV 83
Query 107 SIVIS-SVMGSPDVYSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQPKLFF 165
++++S V + Y+ F AGGL GL L AG AIGI GDAGVRA +QQP++F
Sbjct 84 AVIVSGKVEPAGANYTINNAFSQFAGGLVCGLCGLGAGYAIGIAGDAGVRALSQQPRMFV 143
Query 166 GVTLILIFAEAIGLYGLIIALVI 188
G+ LILIFAE +GLYG+I+AL++
Sbjct 144 GMILILIFAEVLGLYGMIVALIL 166
> Hs20562249
Length=112
Score = 130 bits (327), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 68/112 (60%), Positives = 89/112 (79%), Gaps = 2/112 (1%)
Query 80 VMRPDLIMRSILPVIMAGILGIYGLIISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSS 139
VMRP+ IM+SI+PV+MAGI+ IYGL+++++I++ + D S Y F L GL+VGLS
Sbjct 3 VMRPEQIMKSIIPVVMAGIIAIYGLVVAVLIANSLN--DDISLYKSFLQLGAGLSVGLSG 60
Query 140 LAAGLAIGIVGDAGVRANAQQPKLFFGVTLILIFAEAIGLYGLIIALVIVTR 191
LAAG AIGIVGDAGVR AQQP+LF G+ LILIFAE +GLYGLI+AL++ T+
Sbjct 61 LAAGFAIGIVGDAGVRGTAQQPRLFVGMILILIFAEVLGLYGLIVALILSTK 112
> Hs22060241
Length=138
Score = 125 bits (314), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 70/133 (52%), Positives = 93/133 (69%), Gaps = 19/133 (14%)
Query 59 FANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLIISIVISSVMGSPD 118
F+ AAYGT K+G G++++ VMRP+LIM+SI+PV+ AGI+ IYGL+ +
Sbjct 25 FSAPRAAYGTVKTGAGIAAMSVMRPELIMKSIIPVVTAGIIAIYGLV------------E 72
Query 119 VYSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQPKLFFGVTLILIFAEAIG 178
+ A AG L+VGLS LAAG AI I+GDAGVRA AQQP+LF G+ LILIF E +G
Sbjct 73 LPPASAG-------LSVGLSGLAAGFAIDILGDAGVRATAQQPRLFMGMILILIFPEVLG 125
Query 179 LYGLIIALVIVTR 191
LYGL++AL++ T
Sbjct 126 LYGLVVALILSTE 138
> Hs4757816
Length=205
Score = 82.8 bits (203), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 56/166 (33%), Positives = 93/166 (56%), Gaps = 14/166 (8%)
Query 37 AFLQCDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMA 96
A+ + + + +GI AI + +GAA+G +G + GV P + ++++ +I
Sbjct 38 AWFLTETSPFMWSNLGIGLAISLSVVGAAWGIYITGSSIIGGGVKAPRIKTKNLVSIIFC 97
Query 97 GILGIYGLIISIVISSVMGSPDVYSA-----------YAGFGHLAGGLAVGLSSLAAGLA 145
+ IYG+I++IVIS+ M P +SA +AG+ GL VGLS+L G+
Sbjct 98 EAVAIYGIIMAIVISN-MAEP--FSATDPKAIGHRNYHAGYSMFGAGLTVGLSNLFCGVC 154
Query 146 IGIVGDAGVRANAQQPKLFFGVTLILIFAEAIGLYGLIIALVIVTR 191
+GIVG A+AQ P LF + ++ IF AIGL+G+I+A++ +R
Sbjct 155 VGIVGSGAALADAQNPSLFVKILIVEIFGSAIGLFGVIVAILQTSR 200
> SPAC2C4.13
Length=199
Score = 82.8 bits (203), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 57/182 (31%), Positives = 91/182 (50%), Gaps = 8/182 (4%)
Query 19 VGFPHPEQQPSRARMFWSAFLQCDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSV 78
VG + F S L P + +G +GI S + F +GAA+G G +
Sbjct 18 VGLYMLFHNSGESFDFGSFLLDTSPYT--WGLLGIASCVAFGIIGAAWGIFICGTSILGG 75
Query 79 GVMRPDLIMRSILPVIMAGILGIYGLIISIVISSVMGSPD---VYSA---YAGFGHLAGG 132
V P + ++++ +I ++ IY LII+IV S+ + + Y+ Y GF GG
Sbjct 76 AVKAPRIKTKNLISIIFCEVVAIYSLIIAIVFSAKINDINPAGFYTKSHYYTGFALFWGG 135
Query 133 LAVGLSSLAAGLAIGIVGDAGVRANAQQPKLFFGVTLILIFAEAIGLYGLIIALVIVTRK 192
+ VGL +L G+ +GI G + A+AQ LF V ++ IF +GL+GLI+ L+I +
Sbjct 136 ITVGLCNLICGVCVGITGSSAALADAQDASLFVKVLVVEIFGSVLGLFGLIVGLLIGGKA 195
Query 193 VD 194
D
Sbjct 196 SD 197
> Hs22065586
Length=174
Score = 76.3 bits (186), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 51/134 (38%), Positives = 69/134 (51%), Gaps = 38/134 (28%)
Query 45 SVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLI-MRSILPVIMAGILGIYG 103
++ F G + +V + LGAA G AK+G G+ ++ VM P+LI M+SI+PV+MAGI+ IYG
Sbjct 9 ALVFTISGAMATMVSSGLGAACGMAKNGTGIMAMSVMWPELIHMKSIIPVVMAGIITIYG 68
Query 104 LIISIVISSVMGSPDVYSAYAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQPKL 163
L G AI IVGD G AQQP+L
Sbjct 69 L-------------------------------------PGFAIVIVGDTGKCGTAQQPRL 91
Query 164 FFGVTLILIFAEAI 177
F G+ LILIFA+ +
Sbjct 92 FVGMILILIFAKVL 105
> At4g32530
Length=180
Score = 72.8 bits (177), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 48/165 (29%), Positives = 91/165 (55%), Gaps = 8/165 (4%)
Query 30 RARMFWSAFLQCDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRS 89
A + +A ++ P + F +GI +I + LGAA+G +G + + P + ++
Sbjct 9 HASSWGAALVRISPYT--FSAIGIAISIGVSVLGAAWGIYITGSSLIGAAIEAPRITSKN 66
Query 90 ILPVIMAGILGIYGLIISIVISSVMGSP------DVYSAYAGFGHLAGGLAVGLSSLAAG 143
++ VI + IYG+I++I++ + + S D S AG+ A G+ VG ++L G
Sbjct 67 LISVIFCEAVAIYGVIVAIILQTKLESVPSSKMYDAESLRAGYAIFASGIIVGFANLVCG 126
Query 144 LAIGIVGDAGVRANAQQPKLFFGVTLILIFAEAIGLYGLIIALVI 188
L +GI+G + ++AQ LF + +I IF A+GL+G+I+ +++
Sbjct 127 LCVGIIGSSCALSDAQNSTLFVKILVIEIFGSALGLFGVIVGIIM 171
Score = 35.8 bits (81), Expect = 0.056, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 39/66 (59%), Gaps = 0/66 (0%)
Query 126 FGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQPKLFFGVTLILIFAEAIGLYGLIIA 185
F + +++G+S L A I I G + + A + P++ + +IF EA+ +YG+I+A
Sbjct 25 FSAIGIAISIGVSVLGAAWGIYITGSSLIGAAIEAPRITSKNLISVIFCEAVAIYGVIVA 84
Query 186 LVIVTR 191
+++ T+
Sbjct 85 IILQTK 90
> At2g25610
Length=178
Score = 72.8 bits (177), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 48/165 (29%), Positives = 91/165 (55%), Gaps = 8/165 (4%)
Query 30 RARMFWSAFLQCDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRS 89
A + +A ++ P + F +GI +I + LGAA+G +G + + P + ++
Sbjct 7 HASSWGAALVRISPYT--FSAIGIAISIGVSVLGAAWGIYITGSSLIGAAIEAPRITSKN 64
Query 90 ILPVIMAGILGIYGLIISIVISSVMGSP------DVYSAYAGFGHLAGGLAVGLSSLAAG 143
++ VI + IYG+I++I++ + + S D S AG+ A G+ VG ++L G
Sbjct 65 LISVIFCEAVAIYGVIVAIILQTKLESVPSSKMYDAESLRAGYAIFASGIIVGFANLVCG 124
Query 144 LAIGIVGDAGVRANAQQPKLFFGVTLILIFAEAIGLYGLIIALVI 188
L +GI+G + ++AQ LF + +I IF A+GL+G+I+ +++
Sbjct 125 LCVGIIGSSCALSDAQNSTLFVKILVIEIFGSALGLFGVIVGIIM 169
Score = 36.2 bits (82), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 39/66 (59%), Gaps = 0/66 (0%)
Query 126 FGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQPKLFFGVTLILIFAEAIGLYGLIIA 185
F + +++G+S L A I I G + + A + P++ + +IF EA+ +YG+I+A
Sbjct 23 FSAIGIAISIGVSVLGAAWGIYITGSSLIGAAIEAPRITSKNLISVIFCEAVAIYGVIVA 82
Query 186 LVIVTR 191
+++ T+
Sbjct 83 IILQTK 88
> 7299941
Length=201
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 57/174 (32%), Positives = 89/174 (51%), Gaps = 18/174 (10%)
Query 32 RMFWSAFLQCDPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSIL 91
RM FL N + MGI A + LGAA G G V+ GV P + ++++
Sbjct 27 RMGLGWFLY-TSNPFLWSGMGIFLACALSVLGAASGIYMIGCSVAGGGVRSPRIKTKNLI 85
Query 92 PVIMAGILGIYGLIISIVISSVMGSPDVYSA--------------YAGFGHLAGGLAVGL 137
VI + IYGLI +I++S G+ + +S+ + GF GL VG+
Sbjct 86 SVIFCEAVAIYGLITAILLS---GNVNKFSSVRLITDSTVMATNMFTGFATFGAGLCVGM 142
Query 138 SSLAAGLAIGIVGDAGVRANAQQPKLFFGVTLILIFAEAIGLYGLIIALVIVTR 191
++A G+A+GIVG A+A LF + ++ IF AIGL+GLI+A+ + ++
Sbjct 143 VNVACGIAVGIVGSGAALADAANSALFVKILIVEIFGSAIGLFGLIVAIYMTSK 196
> CE03595
Length=214
Score = 66.2 bits (160), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 59/192 (30%), Positives = 90/192 (46%), Gaps = 31/192 (16%)
Query 10 LKGQRKRFFVGFPHPEQQPSRARMFWSAFLQCDPNSVFFGFMGITSAIVFANLGAAYGTA 69
L GQ RF +G+ P W+ +GI ++ + LGA +G
Sbjct 30 LSGQGHRFDIGWFLTSTSPH----MWAG-------------LGIGFSLSLSVLGAGWGIF 72
Query 70 KSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLIISIVISSVMGS------PDVYSAY 123
+G + GV P + ++++ +I + I+G+I++ V + PD
Sbjct 73 TTGSSILGGGVKAPRIRTKNLVSIIFCEAVAIFGIIMAFVFVGKLAEFRREDLPDTEDGM 132
Query 124 A--------GFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQPKLFFGVTLILIFAE 175
A G+ GGL VGLS+L GLA+GIVG A+A P LF + +I IFA
Sbjct 133 AILARNLASGYMIFGGGLTVGLSNLVCGLAVGIVGSGAAIADAANPALFVKILIIEIFAS 192
Query 176 AIGLYGLIIALV 187
AIGL+G+II +V
Sbjct 193 AIGLFGMIIGIV 204
Score = 31.6 bits (70), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 21/71 (29%), Positives = 37/71 (52%), Gaps = 4/71 (5%)
Query 128 HLAGGLAVG----LSSLAAGLAIGIVGDAGVRANAQQPKLFFGVTLILIFAEAIGLYGLI 183
H+ GL +G LS L AG I G + + + P++ + +IF EA+ ++G+I
Sbjct 49 HMWAGLGIGFSLSLSVLGAGWGIFTTGSSILGGGVKAPRIRTKNLVSIIFCEAVAIFGII 108
Query 184 IALVIVTRKVD 194
+A V V + +
Sbjct 109 MAFVFVGKLAE 119
> 7299943
Length=212
Score = 65.9 bits (159), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 50/165 (30%), Positives = 85/165 (51%), Gaps = 17/165 (10%)
Query 42 DPNSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGI 101
N + +GI ++ + +GAA G +G + GV P + ++++ VI + I
Sbjct 45 SSNPYMWACLGIGLSVSLSVVGAALGIHTTGTSIVGGGVKAPRIKTKNLISVIFCEAVAI 104
Query 102 YGLIISIVIS--------------SVMGSPDVYSAYAGFGHLAGGLAVGLSSLAAGLAIG 147
YGLI +IV+S + + + + +S Y FG GLAVGL +L G+A+G
Sbjct 105 YGLITAIVLSGQLEQFSMETALSQAAIQNTNWFSGYLIFG---AGLAVGLVNLFCGIAVG 161
Query 148 IVGDAGVRANAQQPKLFFGVTLILIFAEAIGLYGLIIALVIVTRK 192
IVG ++A LF + ++ IF AIGL+GLI+ + + ++
Sbjct 162 IVGSGAALSDAANAALFVKILIVEIFGSAIGLFGLIVGIYMTSKS 206
> YHR026w
Length=213
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 42/129 (32%), Positives = 70/129 (54%), Gaps = 6/129 (4%)
Query 51 MGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYGLIISIVI 110
+GI + + +GAA+G +G + GV P + ++++ +I ++ IYGLII+IV
Sbjct 62 LGIALCVGLSVVGAAWGIFITGSSMIGAGVRAPRITTKNLISIIFCEVVAIYGLIIAIVF 121
Query 111 SS---VMGSPDVYSA---YAGFGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQPKLF 164
SS V + ++YS Y G+ G+ VG S+L G+A+GI G ++A LF
Sbjct 122 SSKLTVATAENMYSKSNLYTGYSLFWAGITVGASNLICGIAVGITGATAAISDAADSALF 181
Query 165 FGVTLILIF 173
+ +I IF
Sbjct 182 VKILVIEIF 190
Score = 37.7 bits (86), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 37/66 (56%), Gaps = 0/66 (0%)
Query 126 FGHLAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQPKLFFGVTLILIFAEAIGLYGLIIA 185
+ +L L VGLS + A I I G + + A + P++ + +IF E + +YGLIIA
Sbjct 59 WANLGIALCVGLSVVGAAWGIFITGSSMIGAGVRAPRITTKNLISIIFCEVVAIYGLIIA 118
Query 186 LVIVTR 191
+V ++
Sbjct 119 IVFSSK 124
> AtCh010
Length=81
Score = 36.6 bits (83), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 38/65 (58%), Gaps = 4/65 (6%)
Query 129 LAGGLAVGLSSLAAGLAIGIVGDAGVRANAQQP----KLFFGVTLILIFAEAIGLYGLII 184
+A GLAVGL+S+ G+ G V A+QP K+ + L L F EA+ +YGL++
Sbjct 11 IAAGLAVGLASIGPGVGQGTAAGQAVEGIARQPEAEGKIRGTLLLSLAFMEALTIYGLVV 70
Query 185 ALVIV 189
AL ++
Sbjct 71 ALALL 75
> ECU07g1350
Length=154
Score = 34.3 bits (77), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 24/56 (42%), Positives = 31/56 (55%), Gaps = 6/56 (10%)
Query 136 GLSSLAAGLAIGIVGDAGVRANAQQPK---LFFGVTLILIFAEAIGLYGLIIALVI 188
G+SS AG +IG +QQ K +FF LILIF E +GL GL+ A+ I
Sbjct 95 GVSSGVAGYSIGHSAKVACVTRSQQKKFNSIFF---LILIFGEVVGLLGLVCAMAI 147
> Hs17446006
Length=168
Score = 32.7 bits (73), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 18/24 (75%), Positives = 19/24 (79%), Gaps = 0/24 (0%)
Query 132 GLAVGLSSLAAGLAIGIVGDAGVR 155
GL+VGLSSLAAG AI VG A VR
Sbjct 142 GLSVGLSSLAAGCAIIFVGGASVR 165
> ECU06g0190
Length=173
Score = 32.0 bits (71), Expect = 0.93, Method: Compositional matrix adjust.
Identities = 33/135 (24%), Positives = 61/135 (45%), Gaps = 8/135 (5%)
Query 44 NSVFFGFMGITSAIVFANLGAAYGTAKSGVGVSSVGVMRPDLIMRSILPVIMAGILGIYG 103
++ F GI I ++ G + G G + + P + R++L +++ +
Sbjct 26 DAPFLASFGIVMCIALSSFGTSKGYQAIGRYMIGSSIKAPRVGTRALLGIVICEANFFFC 85
Query 104 LIISIVISSVMGSPDVYSAYAGFGHL-AGGLAVGLSSLAAGLAIGIVGDAGVRANAQQPK 162
L++S ++ + M D +Y G L + G G+ S + LA GI+ A +A+ P
Sbjct 86 LVMSNLLLTKM---DNVKSYGGQCILFSAGFIAGVCSYCSSLASGIICAAITMMDAKDPT 142
Query 163 LFFGVTLILIFAEAI 177
LF+ L+F E I
Sbjct 143 LFYK----LVFLEVI 153
Lambda K H
0.327 0.143 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3551102874
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40