bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2806_orf1
Length=117
Score E
Sequences producing significant alignments: (Bits) Value
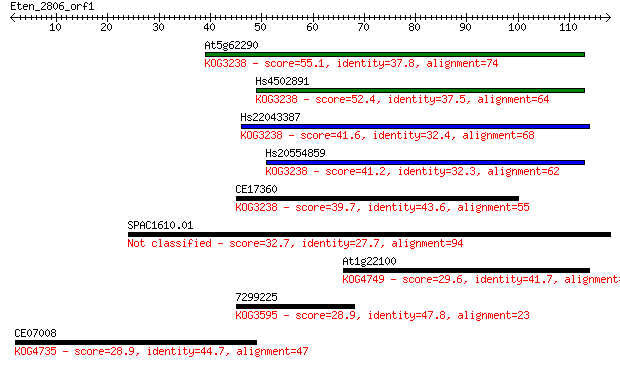
At5g62290 55.1 3e-08
Hs4502891 52.4 2e-07
Hs22043387 41.6 3e-04
Hs20554859 41.2 5e-04
CE17360 39.7 0.001
SPAC1610.01 32.7 0.17
At1g22100 29.6 1.4
7299225 28.9 2.3
CE07008 28.9 2.4
> At5g62290
Length=229
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 28/77 (36%), Positives = 48/77 (62%), Gaps = 9/77 (11%)
Query 39 ELLHNET-IGIKVPNTSVVIRGKLAGCGELFVTSQRVAWLAD--NATSFALNYISIVLHA 95
EL+H +T + + + N + G L++TS+++ WL+D A +A++++SI LHA
Sbjct 28 ELMHVQTSVAVALGNRPI------ESPGTLYITSRKLIWLSDVDMAKGYAVDFLSISLHA 81
Query 96 LSTDPQACDRPCLYCQI 112
+S DP+A PC+Y QI
Sbjct 82 VSRDPEAYSSPCIYTQI 98
> Hs4502891
Length=237
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 35/64 (54%), Gaps = 0/64 (0%)
Query 49 KVPNTSVVIRGKLAGCGELFVTSQRVAWLADNATSFALNYISIVLHALSTDPQACDRPCL 108
+ P+T V+ GK G G L++ R++WL + F+L Y +I LHALS D C L
Sbjct 19 QQPDTEAVLNGKGLGTGTLYIAESRLSWLDGSGLGFSLEYPTISLHALSRDRSDCLGEHL 78
Query 109 YCQI 112
Y +
Sbjct 79 YVMV 82
> Hs22043387
Length=235
Score = 41.6 bits (96), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 33/68 (48%), Gaps = 0/68 (0%)
Query 46 IGIKVPNTSVVIRGKLAGCGELFVTSQRVAWLADNATSFALNYISIVLHALSTDPQACDR 105
I + P+T V+ GK G L++ ++WL + F+L +I LHALS D
Sbjct 24 IDLLQPDTKAVVNGKGLSTGTLYIAESHLSWLDGSGLRFSLECPTICLHALSRDQSDYLG 83
Query 106 PCLYCQIK 113
LY +K
Sbjct 84 EHLYVMVK 91
> Hs20554859
Length=220
Score = 41.2 bits (95), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 31/62 (50%), Gaps = 0/62 (0%)
Query 51 PNTSVVIRGKLAGCGELFVTSQRVAWLADNATSFALNYISIVLHALSTDPQACDRPCLYC 110
P+T V+ K+ G L++ ++WL + F+L Y +I L ALS D C LY
Sbjct 68 PDTKAVLNRKVLRTGTLYIAESHLSWLDSSGLGFSLEYPTISLLALSRDQSDCLGEHLYA 127
Query 111 QI 112
+
Sbjct 128 TV 129
> CE17360
Length=205
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/59 (40%), Positives = 36/59 (61%), Gaps = 4/59 (6%)
Query 45 TIGIKVPNTSVVIRGKLA--GCGELFVTSQRVAWLADNATS--FALNYISIVLHALSTD 99
T GIK+ T+V K+ G G L++T V W++ A + F++ Y +IVLHA+STD
Sbjct 10 TEGIKLATTNVQAFFKIDSLGNGTLYITDSAVIWISSAAGTKGFSVAYPAIVLHAISTD 68
> SPAC1610.01
Length=217
Score = 32.7 bits (73), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 45/96 (46%), Gaps = 9/96 (9%)
Query 24 RMFEHTPHRTPSGDLELLHNETIGIKV-PNTSVVIRGKLAGCGELFVTSQRVAWLADNAT 82
R E+ P+ ++ +T+ K+ + GKL L VTSQ +
Sbjct 14 RKVENGEKLLPNQEIVYFEEKTVPYKIRSDEESFPLGKLIT---LLVTSQSFILFDEEQN 70
Query 83 S-FALNYISIVLHALSTDPQACDRPCLYCQIKGDAL 117
S + + Y +I LHA Q+ D+P +Y Q++G+A+
Sbjct 71 SGWKIPYETITLHA----KQSKDKPYVYVQLEGEAI 102
> At1g22100
Length=457
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 20/51 (39%), Positives = 26/51 (50%), Gaps = 4/51 (7%)
Query 66 ELFVTSQRVAWLADNATSFALNYISIVLH---ALSTDPQACDRPCLYCQIK 113
E +TSQR +W AD A S N S++L L D+PCL +IK
Sbjct 134 EKIITSQRPSWRADVA-SVDTNRSSVLLMDDLTLFAHGHVEDKPCLSVEIK 183
> 7299225
Length=3508
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 11/23 (47%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 45 TIGIKVPNTSVVIRGKLAGCGEL 67
T+ + VP+ ++IR KLA CG L
Sbjct 2084 TVAMMVPDRQIIIRVKLASCGFL 2106
> CE07008
Length=477
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 21/55 (38%), Positives = 26/55 (47%), Gaps = 9/55 (16%)
Query 2 ILRHSAASIPSIGGFCN--------EELGQRMFEHTPHRTPSGDLELLHNETIGI 48
I+ H + IGGF N E++G M H P T SG L NETIG+
Sbjct 364 IIFHKYQNTSQIGGFWNQPKCIIRPEKIGM-MTIHAPMTTYSGLRRSLVNETIGV 417
Lambda K H
0.322 0.137 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171470346
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40