bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2793_orf1
Length=211
Score E
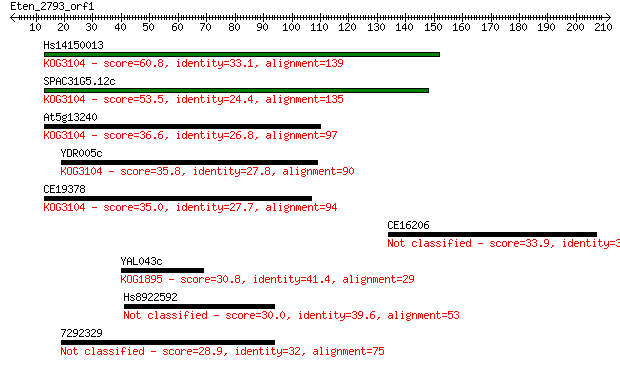
Sequences producing significant alignments: (Bits) Value
Hs14150013 60.8 2e-09
SPAC31G5.12c 53.5 3e-07
At5g13240 36.6 0.036
YDR005c 35.8 0.056
CE19378 35.0 0.11
CE16206 33.9 0.22
YAL043c 30.8 1.9
Hs8922592 30.0 3.3
7292329 28.9 7.0
> Hs14150013
Length=256
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 46/142 (32%), Positives = 66/142 (46%), Gaps = 27/142 (19%)
Query 13 KETLVNLIATLNQCF-PDYEFSSTLTPPMFREEESVAEAQGDINEKL-RAVEQLLPGFIN 70
++TL LIATLN+ F PDY+FS T F E S++ +N L AV +
Sbjct 90 RKTLFYLIATLNESFRPDYDFS-TARSHEFSREPSLSWVVNAVNCSLFSAVREDFKDLKP 148
Query 71 ELWTAIESAVCLSSCAVYSHSA-AGSGEASALGCLWSCLLPPSAAAAAAAAADGGFCLSS 129
+LW A++ +CL+ C +YS++ S G LW S
Sbjct 149 QLWNAVDEEICLAECDIYSYNPDLDSDPFGEDGSLW-----------------------S 185
Query 130 FAYFFVEKQRDRVLFFACCSSS 151
F YFF K+ R++FF+C S S
Sbjct 186 FNYFFYNKRLKRIVFFSCRSIS 207
> SPAC31G5.12c
Length=215
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 33/135 (24%), Positives = 59/135 (43%), Gaps = 24/135 (17%)
Query 13 KETLVNLIATLNQCFPDYEFSSTLTPPMFREEESVAEAQGDINEKLRAVEQLLPGFINEL 72
+ T + ++ATLN +PD++FSS L P F +E S++ +N L + + +N +
Sbjct 62 RRTFMYIVATLNASYPDHDFSS-LQPTDFYKEPSLSRVVDSVNSTLNNIGRGRLS-VNGI 119
Query 73 WTAIESAVCLSSCAVYSHSAAGSGEASALGCLWSCLLPPSAAAAAAAAADGGFCLSSFAY 132
W I+ + LS C+VYS++ + L + +Y
Sbjct 120 WEIIDRHINLSDCSVYSYTPDSDSDPYGDDAL----------------------IWGMSY 157
Query 133 FFVEKQRDRVLFFAC 147
FF K R+L+ +
Sbjct 158 FFFNKNMKRMLYLSL 172
> At5g13240
Length=283
Score = 36.6 bits (83), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 26/104 (25%), Positives = 47/104 (45%), Gaps = 7/104 (6%)
Query 13 KETLVNLIATLNQCFPDYEFSSTLTPPMFREEESVAEAQGDINEKLRAVEQLLP-----G 67
++ L+ L+ TL Q +PDY+FS+ F EE Q N A ++
Sbjct 136 RKALIYLVLTLYQMYPDYDFSAVKAHQFFSEESWDTFKQIFNNYMFEASKEWTERNEDGS 195
Query 68 FINELWTAIESAVCLSSCAVYSHSAAGSGEAS-ALGCLWS-CLL 109
+ ++ A++ V L+ C +Y ++ + + G +WS C L
Sbjct 196 LLEVIYKALDEVVKLAECEIYVYNPNPNADPFLEEGAIWSFCFL 239
> YDR005c
Length=395
Score = 35.8 bits (81), Expect = 0.056, Method: Compositional matrix adjust.
Identities = 25/93 (26%), Positives = 45/93 (48%), Gaps = 11/93 (11%)
Query 19 LIATLNQCFPDYEFSSTLTPPMFRE---EESVAEAQGDINEKLRAVEQLLPGFINELWTA 75
LIA LN +PD++FSS + P F + + +++ + + R E+ + W
Sbjct 238 LIAILNASYPDHDFSS-VEPTDFVKTSLKTFISKFENTLYSLGRQPEEWV-------WEV 289
Query 76 IESAVCLSSCAVYSHSAAGSGEASALGCLWSCL 108
I S + LS C ++ +S + S G LW+ +
Sbjct 290 INSHMTLSDCVLFQYSPSNSFLEDEPGYLWNLI 322
> CE19378
Length=245
Score = 35.0 bits (79), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 26/102 (25%), Positives = 45/102 (44%), Gaps = 17/102 (16%)
Query 13 KETLVNLIATLNQCFPDYEFSSTLTPPMFREEESVAEAQGDINEKLRAVEQLLPGFIN-- 70
++ L +L LN FPD++FS+ S A A + ++ R V+ L +
Sbjct 108 RKKLYDLTQVLNCSFPDHDFSNA---------NSEAFALVNYSDLSRLVDMKLETIVRDY 158
Query 71 -----ELWTAIESAVCLSSCAVYSHSAAGSGEA-SALGCLWS 106
ELW I+ A+ C +YS + + + GC+W+
Sbjct 159 HVRREELWGIIDEAIVPGDCQIYSFKSQFEDDPFTEDGCIWA 200
> CE16206
Length=636
Score = 33.9 bits (76), Expect = 0.22, Method: Composition-based stats.
Identities = 23/73 (31%), Positives = 34/73 (46%), Gaps = 0/73 (0%)
Query 134 FVEKQRDRVLFFACCSSSKLSSPSWLGALDSCQLHDQDDSEEVHLGEVYRSEGLFQGDAE 193
FV +++ F S+S + S G + LH QDD V+ L Q + +
Sbjct 563 FVRMLDEKINFSPEPSTSSDIASSVKGYMSQSFLHQQDDEAPDCTKNVHSESDLKQAEPQ 622
Query 194 EADSLSDLELPSS 206
E+D SD ELPS+
Sbjct 623 ESDKQSDKELPSN 635
> YAL043c
Length=785
Score = 30.8 bits (68), Expect = 1.9, Method: Composition-based stats.
Identities = 12/29 (41%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 40 MFREEESVAEAQGDINEKLRAVEQLLPGF 68
M +EEES E GD N+ A++++ P F
Sbjct 547 MLQEEESAQEISGDANKSTSAIKEIAPPF 575
> Hs8922592
Length=709
Score = 30.0 bits (66), Expect = 3.3, Method: Composition-based stats.
Identities = 21/59 (35%), Positives = 29/59 (49%), Gaps = 16/59 (27%)
Query 41 FREEESVAE------AQGDINEKLRAVEQLLPGFINELWTAIESAVCLSSCAVYSHSAA 93
+ EEE AE A+GDIN+KL+ V Q L +E +C CAV+ + A
Sbjct 11 YMEEEYEAEFQVKITAKGDINQKLQKVIQWL----------LEEKLCALQCAVFDKTLA 59
> 7292329
Length=830
Score = 28.9 bits (63), Expect = 7.0, Method: Composition-based stats.
Identities = 24/82 (29%), Positives = 39/82 (47%), Gaps = 10/82 (12%)
Query 19 LIATLNQCFPDYEFSSTLTPPMFREEESVAEAQ-----GDINEKLRAVEQLLPGFINELW 73
+IA L+Q ++ L FR++ ++ A+ GD + L +LP + E W
Sbjct 444 IIALLSQAMAQFKIYKCLR---FRKKLAIDMAEEYLKSGDHAKALTLYSLMLPDYRQEKW 500
Query 74 TAIESAVCLSS--CAVYSHSAA 93
T I + V L + CA+ S S A
Sbjct 501 TTIFTDVLLKTLRCALLSGSVA 522
Lambda K H
0.317 0.130 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3825978242
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40