bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2775_orf2
Length=319
Score E
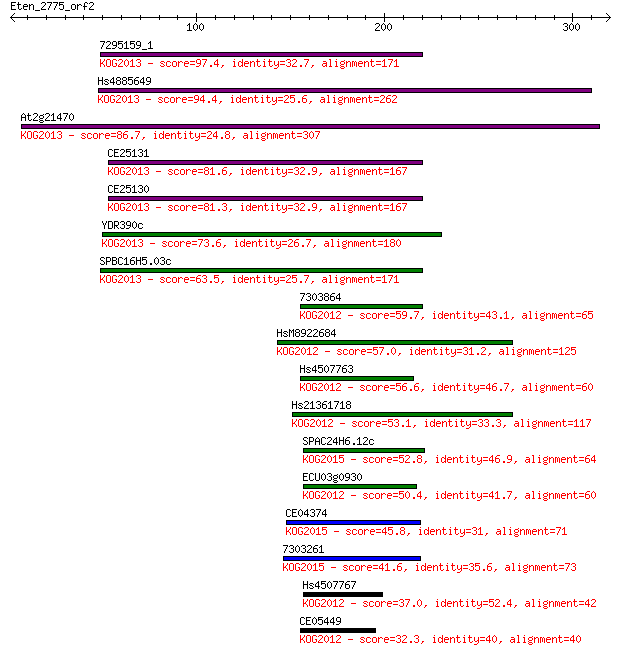
Sequences producing significant alignments: (Bits) Value
7295159_1 97.4 3e-20
Hs4885649 94.4 3e-19
At2g21470 86.7 6e-17
CE25131 81.6 2e-15
CE25130 81.3 3e-15
YDR390c 73.6 5e-13
SPBC16H5.03c 63.5 5e-10
7303864 59.7 8e-09
HsM8922684 57.0 6e-08
Hs4507763 56.6 6e-08
Hs21361718 53.1 8e-07
SPAC24H6.12c 52.8 1e-06
ECU03g0930 50.4 4e-06
CE04374 45.8 1e-04
7303261 41.6 0.002
Hs4507767 37.0 0.052
CE05449 32.3 1.5
> 7295159_1
Length=731
Score = 97.4 bits (241), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 56/171 (32%), Positives = 90/171 (52%), Gaps = 23/171 (13%)
Query 49 RVMEEVFSKQIKDLLKLKKNWKSGKEPQPPLIQPQLLLQWYQQQQQQQQGQQQQQWQQPS 108
++ + F++ I LL++ WK+ K P P +QW + G Q+ +Q
Sbjct 273 KLFNKFFNEDITYLLRMSNLWKTRKAPVP--------VQWDTLLPEGSSGDQKDVAKQH- 323
Query 109 PHKIWSVEECQKIFLRTFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDFV 168
HK+WS+EEC ++F + L +L + + +DKDD AMDFV
Sbjct 324 -HKVWSIEECAQVFANSLKELSANFLKL-------------EGDDTLAWDKDDQPAMDFV 369
Query 169 AAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLL 219
AA + +R + F I K+R+E++++ G IIPAIA+TNAI A + V+ A +L
Sbjct 370 AACANVRSHIFDIERKSRFEIKSMAGNIIPAIATTNAITAGISVMRAFKVL 420
> Hs4885649
Length=640
Score = 94.4 bits (233), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 67/263 (25%), Positives = 130/263 (49%), Gaps = 45/263 (17%)
Query 48 VRVMEEVFSKQIKDLLKLKKNWKSGKEPQPPLIQPQLLLQWYQQQQQQQQGQQQQQWQQP 107
V++ ++F I+ LL + K W+ K P P L W + Q Q ++ Q +P
Sbjct 252 VKLFTKLFKDDIRYLLTMDKLWRKRKPPVP--------LDWAEVQSQGEETNASDQQNEP 303
Query 108 SPHKIWSVEECQKIFLRTFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDF 167
+++ Q + ++++ L + L+ ++ + + +DKDD AMDF
Sbjct 304 Q----LGLKDQQVLDVKSYARLFSKSIETLRVHLAEKGDGAE-----LIWDKDDPSAMDF 354
Query 168 VAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLLSFMEQQKL 227
V +A+ +RM+ F + K+R++++++ G IIPAIA+TNA++A + V+E + +LS Q
Sbjct 355 VTSAANLRMHIFSMNMKSRFDIKSMAGNIIPAIATTNAVIAGLIVLEGLKILSGKIDQ-- 412
Query 228 QKEQQQQQQQPEKTVRDSLARQVWVKPFVTGRRSEEFGLLLLPEALEAPRSSCFVC-QQI 286
R +++ R+ LL+P AL+ P +C+VC +
Sbjct 413 -------------------CRTIFLNKQPNPRKK-----LLVPCALDPPNPNCYVCASKP 448
Query 287 TVTVKLASLKQWSLLAFLTKVLQ 309
VTV+L ++ + ++L K+++
Sbjct 449 EVTVRL-NVHKVTVLTLQDKIVK 470
> At2g21470
Length=700
Score = 86.7 bits (213), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 76/320 (23%), Positives = 137/320 (42%), Gaps = 58/320 (18%)
Query 7 ANLLADLKQDLDALCCCSSSSSSSGSGSG--SGSGGGGLAAAGVRVMEEVFSKQIKDLLK 64
A L D QD D ++S+SSS S + G ++ + VF I+ L
Sbjct 190 AKLFGDKNQDNDLNVRSNNSASSSKETEDVFERSEDEDIEQYGRKIYDHVFGSNIEAALS 249
Query 65 LKKNWKSGKEPQPPLIQPQLLLQWYQQQQQQQQGQQQQQWQ-----QPS-----PHKIWS 114
++ WK+ + P+P + +L + QQ Q PS P ++W
Sbjct 250 NEETWKNRRRPRP-IYSKDVLPESLTQQNGSTQNCSVTDGDLMVSAMPSLGLKNPQELWG 308
Query 115 VEECQKIFLRTFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDFVAAASII 174
+ + +F+ ++++ + + FDKDD LA++FV AA+ I
Sbjct 309 LTQNSLVFIEALKLFFAKRKKEIGH---------------LTFDKDDQLAVEFVTAAANI 353
Query 175 RMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLLSFMEQQKLQKEQQQQ 234
R +F IP + +E + I G I+ A+A+TNAI+A + V+EA+ +L ++ + K +
Sbjct 354 RAESFGIPLHSLFEAKGIAGNIVHAVATTNAIIAGLIVIEAIKVL----KKDVDKFRMTY 409
Query 235 -QQQPEKTVRDSLARQVWVKPFVTGRRSEEFGLLLLPEALEAPRSSCFVCQQITVTVKLA 293
+ P K LLL+P P +C+VC + + +++
Sbjct 410 CLEHPSKK------------------------LLLMPIEPYEPNPACYVCSETPLVLEIN 445
Query 294 SLKQWSLLAFLTKVLQNGLG 313
+ K L + K+++ LG
Sbjct 446 TRKS-KLRDLVDKIVKTKLG 464
> CE25131
Length=438
Score = 81.6 bits (200), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 55/167 (32%), Positives = 87/167 (52%), Gaps = 23/167 (13%)
Query 53 EVFSKQIKDLLKLKKNWKSGKEPQPPLIQPQLLLQWYQQQQQQQQGQQQQQWQQPSPHKI 112
++F I+ L K++ WK K P P L+++ + Q Q+ I
Sbjct 266 QLFLHDIEYLCKMEHLWKQRKRPSP--------LEFHTASSTGGEPQSLCDAQRDDT-SI 316
Query 113 WSVEECQKIFLRTFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDFVAAAS 172
W++ C K+F + C Q LL+Q + + + + FDKD + M FVAA +
Sbjct 317 WTLSTCAKVF--STCI-----QELLEQIRAEPDVK-------LAFDKDHAIIMSFVAACA 362
Query 173 IIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLL 219
IR F IP K++++++A+ G IIPAIASTNAIVA + V EA+ ++
Sbjct 363 NIRAKIFGIPMKSQFDIKAMAGNIIPAIASTNAIVAGIIVTEAVRVI 409
> CE25130
Length=456
Score = 81.3 bits (199), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 55/167 (32%), Positives = 87/167 (52%), Gaps = 23/167 (13%)
Query 53 EVFSKQIKDLLKLKKNWKSGKEPQPPLIQPQLLLQWYQQQQQQQQGQQQQQWQQPSPHKI 112
++F I+ L K++ WK K P P L+++ + Q Q+ I
Sbjct 284 QLFLHDIEYLCKMEHLWKQRKRPSP--------LEFHTASSTGGEPQSLCDAQRDDT-SI 334
Query 113 WSVEECQKIFLRTFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDFVAAAS 172
W++ C K+F + C Q LL+Q + + + + FDKD + M FVAA +
Sbjct 335 WTLSTCAKVF--STCI-----QELLEQIRAEPDVK-------LAFDKDHAIIMSFVAACA 380
Query 173 IIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLL 219
IR F IP K++++++A+ G IIPAIASTNAIVA + V EA+ ++
Sbjct 381 NIRAKIFGIPMKSQFDIKAMAGNIIPAIASTNAIVAGIIVTEAVRVI 427
> YDR390c
Length=636
Score = 73.6 bits (179), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 48/180 (26%), Positives = 86/180 (47%), Gaps = 30/180 (16%)
Query 50 VMEEVFSKQIKDLLKLKKNWKSGKEPQPPLIQPQLLLQWYQQQQQQQQGQQQQQWQQPSP 109
++ ++F + I LL ++ WK+ +P P Q + Q +
Sbjct 253 ILNKLFIQDINKLLAIENLWKTRTKPVP---------------LSDSQINTPTKTAQSAS 297
Query 110 HKIWSVEECQKIFLRTFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDFVA 169
+ + +++E F+ LM+R + +Q I FDKDD ++FVA
Sbjct 298 NSVGTIQEQISNFINITQKLMDRYPK---------------EQNHIEFDKDDADTLEFVA 342
Query 170 AASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLLSFMEQQKLQK 229
A+ IR + F+IP K+ ++++ I G IIPAIA+TNAIVA + ++ +L+ ++ K
Sbjct 343 TAANIRSHIFNIPMKSVFDIKQIAGNIIPAIATTNAIVAGASSLISLRVLNLLKYAPTTK 402
> SPBC16H5.03c
Length=628
Score = 63.5 bits (153), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 44/171 (25%), Positives = 84/171 (49%), Gaps = 27/171 (15%)
Query 49 RVMEEVFSKQIKDLLKLKKNWKSGKEPQPPLIQPQLLLQWYQQQQQQQQGQQQQQWQQPS 108
++ ++F+K I L ++ W + + P L +LL + + W +
Sbjct 255 KIFTKMFTKDIVRLREVPDAW-TYRSPPKELSYSELL---------ENAEKATSPWL--N 302
Query 109 PHKIWSVEECQKIFLRTFCALMERKQRLLQQQQQQQQQQQQQQQQGIPFDKDDDLAMDFV 168
+W+V E +F L + +RL + + + + FDKDD +DFV
Sbjct 303 EQNVWNVAE-------SFAVLRDSIRRL--------ALRSKSSKDDLSFDKDDKDTLDFV 347
Query 169 AAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVEAMHLL 219
AAA+ +R + F I + ++++ + G IIPAIA+TNA++A + + +A+ +L
Sbjct 348 AAAANLRAHVFGIQQLSEFDIKQMAGNIIPAIATTNAVIAGLCITQAIKVL 398
> 7303864
Length=1008
Score = 59.7 bits (143), Expect = 8e-09, Method: Composition-based stats.
Identities = 28/67 (41%), Positives = 45/67 (67%), Gaps = 2/67 (2%)
Query 155 IPFDKDDD--LAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQV 212
+ F+KDDD L MDF+ A S +R + IPP +R + + I G IIPAIA+T ++++ + V
Sbjct 803 LEFEKDDDSNLHMDFIVACSNLRAANYKIPPADRHKSKLIAGKIIPAIATTTSVLSGLAV 862
Query 213 VEAMHLL 219
+E + L+
Sbjct 863 LEVIKLI 869
> HsM8922684
Length=459
Score = 57.0 bits (136), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 39/147 (26%), Positives = 70/147 (47%), Gaps = 34/147 (23%)
Query 143 QQQQQQQQQQQGIPFDKDDDL--AMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAI 200
+ + Q + F+KDDD +DF+ AAS +R + I P +R++ + I G IIPAI
Sbjct 242 NEATKSDLQMAVLSFEKDDDHNGHIDFITAASNLRAKMYSIEPADRFKTKRIAGKIIPAI 301
Query 201 ASTNAIVAAMQVVE--------------------AMHLLSFMEQQKLQKEQQQQQQQPEK 240
A+T A V+ + +E A+ ++ F E +++K +
Sbjct 302 ATTTATVSGLVALEMIKVTGGYPFEAYKNCFLNLAIPIVVFTETTEVRKTK--------- 352
Query 241 TVRDSLARQVWVKPFVTGRRSEEFGLL 267
+R+ ++ +W + V G+ E+F LL
Sbjct 353 -IRNGISFTIWDRWTVHGK--EDFTLL 376
> Hs4507763
Length=1058
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 38/62 (61%), Gaps = 2/62 (3%)
Query 155 IPFDKDDD--LAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQV 212
I F+KDDD MDF+ AAS +R + IP +R + + I G IIPAIA+T A V +
Sbjct 847 IDFEKDDDSNFHMDFIVAASNLRAENYDIPSADRHKSKLIAGKIIPAIATTTAAVVGLVC 906
Query 213 VE 214
+E
Sbjct 907 LE 908
> Hs21361718
Length=1052
Score = 53.1 bits (126), Expect = 8e-07, Method: Composition-based stats.
Identities = 39/139 (28%), Positives = 68/139 (48%), Gaps = 34/139 (24%)
Query 151 QQQGIPFDKDDDLA--MDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVA 208
Q + F+KDDD +DF+ AAS +R + I P +R++ + I G IIPAIA+T A V+
Sbjct 843 QMAVLSFEKDDDHNGHIDFITAASNLRAKMYSIEPADRFKTKRIAGKIIPAIATTTATVS 902
Query 209 AMQVVE--------------------AMHLLSFMEQQKLQKEQQQQQQQPEKTVRDSLAR 248
+ +E A+ ++ F E +++K + +R+ ++
Sbjct 903 GLVALEMIKVTGGYPFEVYKNCFLNLAIPIVVFTETTEVRKTK----------IRNGISF 952
Query 249 QVWVKPFVTGRRSEEFGLL 267
+W + V G+ E+F LL
Sbjct 953 TIWDRWTVHGK--EDFTLL 969
> SPAC24H6.12c
Length=444
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/66 (45%), Positives = 42/66 (63%), Gaps = 2/66 (3%)
Query 157 FDKDDDLAMDFVAAASIIRMNTFHIPPK--NRWELQAIVGAIIPAIASTNAIVAAMQVVE 214
F+ D+ +D++ SI R N F IP NR+ +Q IV IIPA+ASTNAI+AA E
Sbjct 254 FEPDNIRHIDWLVKRSIERANKFQIPSSSINRFFVQGIVKRIIPAVASTNAIIAASCCNE 313
Query 215 AMHLLS 220
A+ +L+
Sbjct 314 ALKILT 319
> ECU03g0930
Length=991
Score = 50.4 bits (119), Expect = 4e-06, Method: Composition-based stats.
Identities = 25/62 (40%), Positives = 40/62 (64%), Gaps = 2/62 (3%)
Query 157 FDKDDD--LAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIVAAMQVVE 214
F+KDDD +DF+ AA+ +R + I +R ++ I G IIPAIA+T A+V+ + V+E
Sbjct 797 FEKDDDTNFHVDFLYAAANLRAINYKIKQADRLTVKGIAGRIIPAIATTTAVVSGLAVLE 856
Query 215 AM 216
+
Sbjct 857 MI 858
> CE04374
Length=437
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 43/71 (60%), Gaps = 0/71 (0%)
Query 148 QQQQQQGIPFDKDDDLAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNAIV 207
+++ +G+ D DD + +++V + +R ++I +R ++ IIPA+ASTNA++
Sbjct 241 EEKPFEGVSLDADDPIHVEWVLERASLRAEKYNIRGVDRRLTSGVLKRIIPAVASTNAVI 300
Query 208 AAMQVVEAMHL 218
AA +EA+ L
Sbjct 301 AASCALEALKL 311
> 7303261
Length=450
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 40/73 (54%), Gaps = 0/73 (0%)
Query 146 QQQQQQQQGIPFDKDDDLAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIPAIASTNA 205
Q ++Q G+P D DD + ++ ++ R N F+I +Q +V IIPA+ASTNA
Sbjct 237 QWEKQNPFGVPLDGDDPQHIGWIYERALERSNEFNITGVTYRLVQGVVKHIIPAVASTNA 296
Query 206 IVAAMQVVEAMHL 218
+AA +E L
Sbjct 297 AIAAACALEVFKL 309
> Hs4507767
Length=1011
Score = 37.0 bits (84), Expect = 0.052, Method: Compositional matrix adjust.
Identities = 22/44 (50%), Positives = 29/44 (65%), Gaps = 2/44 (4%)
Query 157 FDKDDD--LAMDFVAAASIIRMNTFHIPPKNRWELQAIVGAIIP 198
F+KDDD +DFV AA+ +R + IPP NR + + IVG IIP
Sbjct 807 FEKDDDSNFHVDFVVAAASLRCQNYGIPPVNRAQSKRIVGQIIP 850
> CE05449
Length=1113
Score = 32.3 bits (72), Expect = 1.5, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 24/42 (57%), Gaps = 2/42 (4%)
Query 155 IPFDKDDDLA--MDFVAAASIIRMNTFHIPPKNRWELQAIVG 194
+ F+KDDD M+F+ AAS +R + I P +R + I G
Sbjct 904 VDFEKDDDSNHHMEFITAASNLRAENYDILPADRMRTKQIAG 945
Lambda K H
0.318 0.129 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7449381586
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40