bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2760_orf1
Length=180
Score E
Sequences producing significant alignments: (Bits) Value
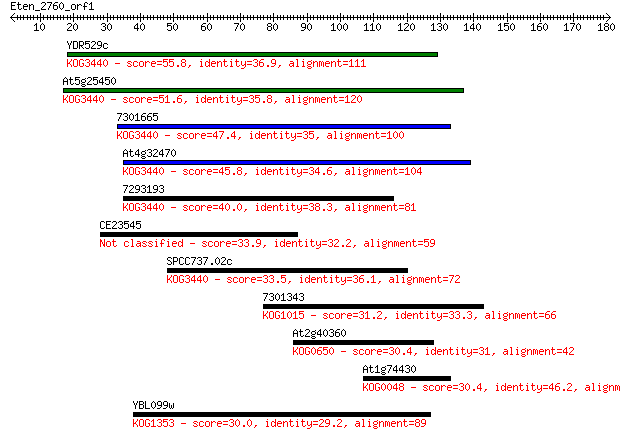
YDR529c 55.8 4e-08
At5g25450 51.6 8e-07
7301665 47.4 2e-05
At4g32470 45.8 5e-05
7293193 40.0 0.002
CE23545 33.9 0.16
SPCC737.02c 33.5 0.23
7301343 31.2 1.1
At2g40360 30.4 2.1
At1g74430 30.4 2.1
YBL099w 30.0 2.6
> YDR529c
Length=127
Score = 55.8 bits (133), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 41/112 (36%), Positives = 58/112 (51%), Gaps = 12/112 (10%)
Query 18 FLSELLAPVWFRFFRGPLDRWNQAAIGKYLREQGLMYDDLYSDKEPVIERALELLPEDLA 77
LS+L PV +F N A K GL +DDL +++ P+++ AL LPED +
Sbjct 20 VLSKLCVPVANQFI-------NLAGYKKL----GLKFDDLIAEENPIMQTALRRLPEDES 68
Query 78 TARYRRIMRATHLNHVRLYLPPNE-QNYDPFIPYLAPYVEEAKFQLQEEEEL 128
AR RI+RA LP NE +PYL PY+ EA+ +E++EL
Sbjct 69 YARAYRIIRAHQTELTHHLLPRNEWIKAQEDVPYLLPYILEAEAAAKEKDEL 120
> At5g25450
Length=122
Score = 51.6 bits (122), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 43/125 (34%), Positives = 61/125 (48%), Gaps = 13/125 (10%)
Query 17 SFLSELLAPVWFRFFRGPLDRWNQAAIGKYLREQGLMYDDLYSDKEPV----IERALELL 72
SFL L+ P + L R + ++ LR GL YDDLY +P+ I+ AL L
Sbjct 3 SFLQRLVDPR-----KNFLARMHMKSVSNRLRRYGLRYDDLY---DPLYDLDIKEALNRL 54
Query 73 PEDLATARYRRIMRATHLNHVRLYLPPNEQNYD-PFIPYLAPYVEEAKFQLQEEEELLGY 131
P ++ AR +R+MRA L+ YLP N Q PF YL + K + E E L
Sbjct 55 PREIVDARNQRLMRAMDLSMKHEYLPDNLQAVQTPFRSYLQDMLALVKRERAEREALGAL 114
Query 132 HMWER 136
+++R
Sbjct 115 PLYQR 119
> 7301665
Length=111
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 35/103 (33%), Positives = 49/103 (47%), Gaps = 8/103 (7%)
Query 33 GPLDRW--NQAAIGKYLREQGLMYDDLYSDKEPVIERALELLPEDLATARYRRIMRATHL 90
L +W N + +Y GL DD + E V E A+ LP L R RI+RA HL
Sbjct 14 SKLGKWAYNMSGFNQY----GLYRDDCLYENEDVAE-AVRRLPRKLYDERNYRILRALHL 68
Query 91 NHVRLYLPPNE-QNYDPFIPYLAPYVEEAKFQLQEEEELLGYH 132
+ + LP + Y+ I YL PY+ E + + +E EE H
Sbjct 69 SMTKTILPKEQWTKYEEDIKYLEPYLNEVQKEREEREEWSKTH 111
> At4g32470
Length=122
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 36/106 (33%), Positives = 53/106 (50%), Gaps = 2/106 (1%)
Query 35 LDRWNQAAIGKYLREQGLMYDDLYSDKEPV-IERALELLPEDLATARYRRIMRATHLNHV 93
L R + AI LR GL YDDLY + I+ A+ LP ++ AR +R+ RA L+
Sbjct 16 LARMHMKAISTRLRRYGLRYDDLYDQYYSMDIKEAMNRLPREVVDARNQRLKRAMDLSMK 75
Query 94 RLYLPPNEQNYD-PFIPYLAPYVEEAKFQLQEEEELLGYHMWERRL 138
YLP + Q PF YL + + + +E E L +++R L
Sbjct 76 HEYLPKDLQAVQTPFRGYLQDMLALVERESKEREALGALPLYQRTL 121
> 7293193
Length=111
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 31/84 (36%), Positives = 41/84 (48%), Gaps = 8/84 (9%)
Query 35 LDRW--NQAAIGKYLREQGLMYDDLYSDKEPVIERALELLPEDLATARYRRIMRATHLNH 92
L RW N + +Y GL DD + E V E A+ LP L R RIMRA HL+
Sbjct 16 LGRWAYNLSGFNQY----GLHRDDCLYENEDVKE-AVRRLPRKLYDERNYRIMRALHLSM 70
Query 93 VRLYLPPNE-QNYDPFIPYLAPYV 115
+ LP + Y+ + YL PY+
Sbjct 71 TKTILPKEQWTKYEEDVKYLEPYL 94
> CE23545
Length=801
Score = 33.9 bits (76), Expect = 0.16, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 32/59 (54%), Gaps = 7/59 (11%)
Query 28 FRFFRGPLDRWNQAAIGKYLREQGLMYDDLYSDKEPVIERALELLPEDLATARYRRIMR 86
F+ +G ++R+ Q GK L DD +K +IE+ALE+L E+ Y+++ R
Sbjct 420 FKEVKGLVERFEQRLKGKEL-------DDEQKNKAKLIEKALEMLLENFDDTDYKKVER 471
> SPCC737.02c
Length=137
Score = 33.5 bits (75), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 38/77 (49%), Gaps = 5/77 (6%)
Query 48 REQGLMYDDLYSDKEPVIERALELLPEDLATARYRRIMRATHLNHVRLYLPPN-----EQ 102
R+ GL YDDL ++ ++AL LP+ + R RI RA L+ LP + E+
Sbjct 34 RKYGLRYDDLMLEENDDTQKALSRLPKMESYDRVYRIRRAMQLSIENKILPKSEWTKPEE 93
Query 103 NYDPFIPYLAPYVEEAK 119
+Y P LA + E K
Sbjct 94 DYHYLRPVLAEVIAERK 110
> 7301343
Length=1311
Score = 31.2 bits (69), Expect = 1.1, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 33/69 (47%), Gaps = 3/69 (4%)
Query 77 ATARYRRIMR-ATHLNHVRLYLPPNEQNYDPFIPYLAPYVEEAKFQLQEE--EELLGYHM 133
+T R R+M+ +H+ H L ++Y PYL P E + E ++L GY+M
Sbjct 688 STERDLRLMKHRSHILHKLLEGCIQRRDYSVLAPYLPPKHEYVVYTTLSELQQKLYGYYM 747
Query 134 WERRLYSGG 142
R SGG
Sbjct 748 TTHREQSGG 756
> At2g40360
Length=775
Score = 30.4 bits (67), Expect = 2.1, Method: Composition-based stats.
Identities = 13/45 (28%), Positives = 29/45 (64%), Gaps = 3/45 (6%)
Query 86 RATHLNHV---RLYLPPNEQNYDPFIPYLAPYVEEAKFQLQEEEE 127
++ HL ++ +L LP ++++Y+P + Y+ E+A ++L EE+
Sbjct 326 KSKHLTYIPPPKLKLPGHDESYNPSLEYIPTEEEKASYELMFEED 370
> At1g74430
Length=271
Score = 30.4 bits (67), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 107 FIPYLAPYVEEAKFQLQEEEELLGYH 132
++ YL P ++ KF QEEEE++ YH
Sbjct 57 WLNYLRPGIKRGKFTPQEEEEIIKYH 82
> YBL099w
Length=545
Score = 30.0 bits (66), Expect = 2.6, Method: Composition-based stats.
Identities = 26/93 (27%), Positives = 39/93 (41%), Gaps = 28/93 (30%)
Query 38 WNQAAIGKYLREQG----LMYDDLYSDKEPVIERALELLPEDLATARYRRIMRATHLNHV 93
+ A+IG++ R+ G ++YDDL K+ V R L LL
Sbjct 285 FTAASIGEWFRDNGKHALIVYDDL--SKQAVAYRQLSLL--------------------- 321
Query 94 RLYLPPNEQNYDPFIPYLAPYVEEAKFQLQEEE 126
L PP + Y + YL P + E +L E+E
Sbjct 322 -LRRPPGREAYPGDVFYLHPRLLERAAKLSEKE 353
Lambda K H
0.322 0.141 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2806646388
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40