bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2744_orf2
Length=481
Score E
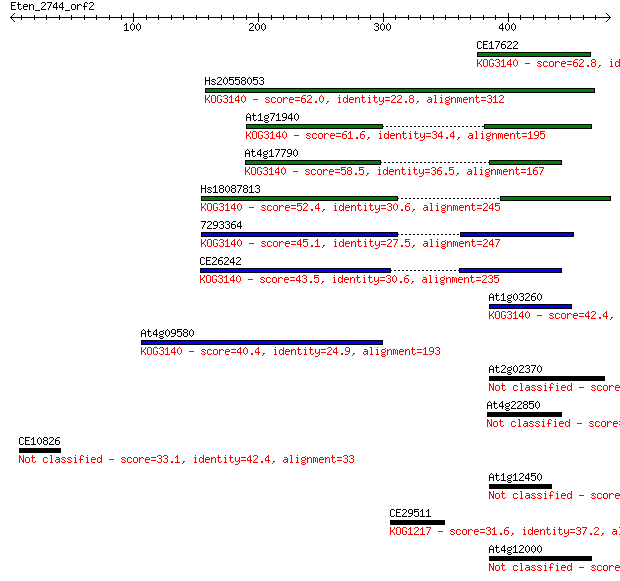
Sequences producing significant alignments: (Bits) Value
CE17622 62.8 1e-09
Hs20558053 62.0 3e-09
At1g71940 61.6 3e-09
At4g17790 58.5 3e-08
Hs18087813 52.4 2e-06
7293364 45.1 3e-04
CE26242 43.5 0.001
At1g03260 42.4 0.002
At4g09580 40.4 0.008
At2g02370 35.8 0.20
At4g22850 34.3 0.65
CE10826 33.1 1.3
At1g12450 31.6 3.6
CE29511 31.6 4.5
At4g12000 30.4 8.8
> CE17622
Length=246
Score = 62.8 bits (151), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 39/102 (38%), Positives = 55/102 (53%), Gaps = 11/102 (10%)
Query 375 WDTEL-----DLMLTVLFLRVSPF-PNLVINAASPVLDVPFGAFFVATWLGLMPNSALFV 428
W +L D + ++FLRV+P PN +IN ASPVLDVP FF T+LG+ P S L++
Sbjct 143 WQDDLSKHRDDFLNYMIFLRVTPIVPNWLINIASPVLDVPLAPFFWGTFLGVAPPSFLYI 202
Query 429 SMGSALGSLDSLSSGWRPLAVFVFGAFAV-----VLLKTALR 465
GS L L S W ++ + A+ +LLK L+
Sbjct 203 QAGSTLEQLSHTSVAWSWSSIVLLTGSAILSLAPILLKKKLK 244
> Hs20558053
Length=291
Score = 62.0 bits (149), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 71/317 (22%), Positives = 123/317 (38%), Gaps = 94/317 (29%)
Query 157 LFVCALWAMAILYLSLPGLTEEARAELESVFPTSFSDLGCLRDIKTLRKLCAAIGVYRQQ 216
+F+ A + M ++Y + P L+EE R ++ RD+ + L + Y+
Sbjct 60 IFLSAAFVMFLVYKNFPQLSEEERVNMK-----------VPRDMDDAKALGKVLSKYKDT 108
Query 217 H--PVALALLLSYVYLLYQAFPLFMFPFSGLAMAVTLLLGALYSPLVAFSLASALSAIGP 274
V +A +Y++L A P +F +++L G LY +A L S +G
Sbjct 109 FYVQVLVAYFATYIFLQTFAIPGSIF--------LSILSGFLYPFPLALFLVCLCSGLGA 160
Query 275 SLSYFLFKAAGKPLVVRLFPNQLQRMRQLIHPTESSSACGSGCSDISSSSNIASNDKSSS 334
S Y L G+P+V
Sbjct 161 SFCYMLSYLVGRPVV--------------------------------------------- 175
Query 335 AAVQLKQKTQTSAEPQGQHQQQREHRQQHLQPRQHRPRCAWDTELDLMLTVLFLRVSPF- 393
K T+ + + Q ++ REH L+ ++FLR++PF
Sbjct 176 ----YKYLTEKAVKWSQQVERHREH---------------------LINYIIFLRITPFL 210
Query 394 PNLVINAASPVLDVPFGAFFVATWLGLMPNSALFVSMGSALGSLDSLSSGWRPLAVFVFG 453
PN IN SPV++VP FF+ T+LG+ P S + + G+ L L + ++F+
Sbjct 211 PNWFINITSPVINVPLKVFFIGTFLGVAPPSFVAIKAGTTLYQLTTAGEAVSWNSIFILM 270
Query 454 AFAVVLLKTAL--RKLR 468
AV+ + A+ +KL+
Sbjct 271 ILAVLSILPAIFQKKLK 287
> At1g71940
Length=272
Score = 61.6 bits (148), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 37/89 (41%), Positives = 54/89 (60%), Gaps = 3/89 (3%)
Query 381 LMLTVLFLRVSP-FPNLVINAASPVLDVPFGAFFVATWLGLMPNSALFVSMGSALGSLDS 439
L+ +LFLR++P PNL IN ASP++DVPF FF+AT +GL+P + + V G A+G L S
Sbjct 180 LLNYMLFLRITPTLPNLFINLASPIVDVPFHVFFLATLIGLIPAAYITVRAGLAIGDLKS 239
Query 440 LSS--GWRPLAVFVFGAFAVVLLKTALRK 466
+ ++ L+V F +L RK
Sbjct 240 VKDLYDFKTLSVLFLIGFISILPTILKRK 268
Score = 38.1 bits (87), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 30/109 (27%), Positives = 49/109 (44%), Gaps = 6/109 (5%)
Query 190 SFSDLGCLRDIKTLRKLCAAIGVYRQQHPVALALLLSYVYLLYQAFPLFMFPFSGLAMAV 249
F L R + LR L + Y ++P L Y+ Q F M P + + +
Sbjct 65 EFGKLKLPRSLADLRLLKDNLANYANEYPAQFVLGYCATYIFMQTF---MIPGT---IFM 118
Query 250 TLLLGALYSPLVAFSLASALSAIGPSLSYFLFKAAGKPLVVRLFPNQLQ 298
+LL GAL+ L + G + +FL K G+PL+ L+P++L+
Sbjct 119 SLLAGALFGVFKGVVLVVFNATAGATSCFFLSKLIGRPLITWLWPDKLR 167
> At4g17790
Length=264
Score = 58.5 bits (140), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 30/59 (50%), Positives = 42/59 (71%), Gaps = 1/59 (1%)
Query 385 VLFLRVSP-FPNLVINAASPVLDVPFGAFFVATWLGLMPNSALFVSMGSALGSLDSLSS 442
+LFLR++P PN IN ASP++DVP+ FF+AT++GL+P + + V G ALG L SL
Sbjct 178 MLFLRLTPTLPNTFINVASPIVDVPYHIFFLATFIGLIPAAFVTVRAGLALGELKSLGD 236
Score = 40.0 bits (92), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 31/109 (28%), Positives = 57/109 (52%), Gaps = 6/109 (5%)
Query 189 TSFSDLGCLRDIKTLRKLCAAIGVYRQQHPVALALLLSYVYLLYQAFPLFMFPFSGLAMA 248
+ +S L R+++ L+ L + +Y + V + + VY+ Q F M P + +
Sbjct 58 SDYSFLKLPRNLEDLQILRDNLEIYTSDYTVQVLVGYCLVYVFMQTF---MIPGT---VF 111
Query 249 VTLLLGALYSPLVAFSLASALSAIGPSLSYFLFKAAGKPLVVRLFPNQL 297
++LL GAL+ + +L + + G S YFL K G+PL+ L+P++L
Sbjct 112 MSLLAGALFGVVKGMALVVSTATAGASSCYFLSKLIGRPLLFSLWPDKL 160
> Hs18087813
Length=264
Score = 52.4 bits (124), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 46/162 (28%), Positives = 75/162 (46%), Gaps = 21/162 (12%)
Query 154 MALLFVCALWAMAILYLSLP-----GLTEEARAELESVFPTSFSDLGCLRDIKTLRKLCA 208
+ L+F +A+ +L LP G TEEA FP+ D+ LR+L
Sbjct 7 LLLVFAGCTFALYLLSTRLPRGRRLGSTEEAGGR-SLWFPS---------DLAELRELSE 56
Query 209 AIGVYRQQHPVALALLLSYVYLLYQAFPLFMFPFSGLAMAVTLLLGALYSPLVAFSLASA 268
+ YR++H + LL YL Q F + P S + +L GAL+ P + L
Sbjct 57 VLREYRKEHQAYVFLLFCGAYLYKQGFAI---PGSSF---LNVLAGALFGPWLGLLLCCV 110
Query 269 LSAIGPSLSYFLFKAAGKPLVVRLFPNQLQRMRQLIHPTESS 310
L+++G + Y L GK LVV FP+++ +++ + +S
Sbjct 111 LTSVGATCCYLLSSIFGKQLVVSYFPDKVALLQRKVEENRNS 152
Score = 47.8 bits (112), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 48/90 (53%), Gaps = 2/90 (2%)
Query 394 PNLVINAASPVLDVPFGAFFVATWLGLMPNSALFVSMGSALGSLDSLSSGWRPLAVFVFG 453
PN +N ++P+L++P FF + +GL+P + + V GS L +L SL + + VF
Sbjct 167 PNWFLNLSAPILNIPIVQFFFSVLIGLIPYNFICVQTGSILSTLTSLDALFSWDTVFKLL 226
Query 454 AFAVVLL--KTALRKLRPCHLAEQDAPTVS 481
A A+V L T ++K HL + T +
Sbjct 227 AIAMVALIPGTLIKKFSQKHLQLNETSTAN 256
> 7293364
Length=304
Score = 45.1 bits (105), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/98 (29%), Positives = 47/98 (47%), Gaps = 11/98 (11%)
Query 362 QHLQPRQHRPRCAWDTELD-----LMLTVLFLRVSPF-PNLVINAASPVLDVPFGAFFVA 415
+H P++ W ++ L +LFLR++P PN IN ASPV+ VP F +
Sbjct 188 RHFWPKK---TSEWSKHVEEYRDSLFNYMLFLRMTPILPNWFINLASPVIGVPLHIFALG 244
Query 416 TWLGLMPNSALFVSMGSALGSLDSLSSG--WRPLAVFV 451
T+ G+ P S + + G L + S S W + + +
Sbjct 245 TFCGVAPPSVIAIQAGKTLQKMTSSSEAFSWTSMGILM 282
Score = 40.4 bits (93), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 39/159 (24%), Positives = 68/159 (42%), Gaps = 21/159 (13%)
Query 154 MALLFVCALWAMAILYLSLPGLTEEARAELESVFPTSFSDLGCLRDIKTLRKLCAAIGVY 213
+A +FV +L M +Y P L + L+ P RDI+ + L + Y
Sbjct 69 VAGIFVASLVTMCYVYAIFPELNASEKQHLK--IP---------RDIQDAKMLAKVLDRY 117
Query 214 RQQH--PVALALLLSYVYLLYQAFPLFMFPFSGLAMAVTLLLGALYSPLVAFSLASALSA 271
+ + V ++++YV+L A P +F +++LLG LY +A L SA
Sbjct 118 KDMYYFEVMFGVVVAYVFLQTFAIPGSLF--------LSILLGFLYKFPIALFLICFCSA 169
Query 272 IGPSLSYFLFKAAGKPLVVRLFPNQLQRMRQLIHPTESS 310
+G +L Y L G+ L+ +P + + + S
Sbjct 170 LGATLCYTLSNLVGRRLIRHFWPKKTSEWSKHVEEYRDS 208
> CE26242
Length=270
Score = 43.5 bits (101), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/84 (33%), Positives = 45/84 (53%), Gaps = 2/84 (2%)
Query 361 QQHLQPRQHRPRCAWDTELD-LMLTVLFLRVSPF-PNLVINAASPVLDVPFGAFFVATWL 418
+ L+ R RC + E D L +L R+ PF P+ ++N +SP LDVP + ++
Sbjct 141 DRFLKSRIESLRCLVNAERDRLWFFLLSARIFPFTPHWLLNISSPFLDVPLRYHASSVFV 200
Query 419 GLMPNSALFVSMGSALGSLDSLSS 442
GL P + L V G+ L ++S+S
Sbjct 201 GLFPYNLLCVRAGTVLAEVNSMSD 224
Score = 43.5 bits (101), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 44/155 (28%), Positives = 73/155 (47%), Gaps = 12/155 (7%)
Query 153 FMALLFVCALWAMAILYLSLPGLTEEAR--AELESVFPTSFSDLGCLRDIKTLRKLCAAI 210
F A L + A+W+ Y LP + R +LE + S S R++K ++ A
Sbjct 12 FGASLVLYAVWS----YGPLPEGAQRPRFPRDLEGLRELSSSLTKYDRNLKKPQRDRNAS 67
Query 211 GVYRQQHPVALALLLSYVYLLYQAFPLFMFPFSGLAMAVTLLLGALYSPLVAFSLASALS 270
+R+ H LL S YL Q F + P S + LL GAL+ + +L +L+
Sbjct 68 SFFRESHAAYTVLLFSAAYLYKQTFAI---PGS---FFMNLLAGALFGTVRGVALVCSLN 121
Query 271 AIGPSLSYFLFKAAGKPLVVRLFPNQLQRMRQLIH 305
A+G SL + L P+V R ++++ +R L++
Sbjct 122 AVGASLCFCLSALFAAPIVDRFLKSRIESLRCLVN 156
> At1g03260
Length=269
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/68 (38%), Positives = 38/68 (55%), Gaps = 2/68 (2%)
Query 385 VLFLRVSPF-PNLVINAASPVLDVPFGAFFVATWLGLM-PNSALFVSMGSALGSLDSLSS 442
VL LRV P P ++N V V G + +ATWLG+M P + V +G+ L L ++
Sbjct 131 VLLLRVVPILPFNMLNYLLSVTPVRLGEYMLATWLGMMQPITFALVYVGTTLKDLSDITH 190
Query 443 GWRPLAVF 450
GW ++VF
Sbjct 191 GWHEVSVF 198
> At4g09580
Length=194
Score = 40.4 bits (93), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 48/195 (24%), Positives = 77/195 (39%), Gaps = 24/195 (12%)
Query 106 EGSMGSVQHGEEALVGHLPAQGL--DGPREGARVDFAPERGRVIFIKFGFMALLFVCALW 163
+G + EE+ A+GL D G R ER + +F +F+
Sbjct 10 DGGARQLVKDEESPAASSAAKGLLNDDSPTGKRTK--SERFPLSRWEFAVFFTVFLVFTT 67
Query 164 AMAILYLSLPGLTEEARAELESVFPTSFSDLGCLRDIKTLRKLCAAIGVYRQQHPVALAL 223
+ +YL++P + L R I LR L +G Y ++ L
Sbjct 68 GLFCIYLTMPA--------------AEYGKLKVPRTISDLRLLKENLGSYASEYQARFIL 113
Query 224 LLSYVYLLYQAFPLFMFPFSGLAMAVTLLLGALYSPLVAFSLASALSAIGPSLSYFLFKA 283
Y+ Q F M P + + ++LL GAL+ + F L + G +FL K
Sbjct 114 GYCSTYIFMQTF---MIPGT---IFMSLLAGALFGVVRGFVLVVLNATAGACSCFFLSKL 167
Query 284 AGKPLVVRLFPNQLQ 298
G+PLV L+P +L+
Sbjct 168 VGRPLVNWLWPEKLR 182
> At2g02370
Length=160
Score = 35.8 bits (81), Expect = 0.20, Method: Composition-based stats.
Identities = 25/105 (23%), Positives = 46/105 (43%), Gaps = 13/105 (12%)
Query 385 VLFLRVSPFPNLVINAASPVLDVPFGAFFVATWLGLMPNSALFVSMGSALGSLDSLSSGW 444
V RVSPFP + N A V + F +F + G++P + +++ G + + + G
Sbjct 30 VAIFRVSPFPYTIFNYAIVVTSMRFWPYFFGSIAGMIPEAFIYIYSGRLIRTFADVQYGH 89
Query 445 RPLAV--FVFGAFAVVL-----------LKTALRKLRPCHLAEQD 476
+ L V+ ++V+ K ALR+L+ E +
Sbjct 90 QRLTTVEIVYNVISLVIAVVTTVAFTVYAKRALRELQNAEANEDE 134
> At4g22850
Length=217
Score = 34.3 bits (77), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 31/60 (51%), Gaps = 0/60 (0%)
Query 383 LTVLFLRVSPFPNLVINAASPVLDVPFGAFFVATWLGLMPNSALFVSMGSALGSLDSLSS 442
L V +R+SPFP ++ N S V +G + + LG++P + + G + +L SS
Sbjct 110 LLVTLIRISPFPYILYNYCSVATRVKYGPYITGSLLGMVPEVFVAIYTGILVRTLAEASS 169
> CE10826
Length=779
Score = 33.1 bits (74), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 17/35 (48%), Gaps = 2/35 (5%)
Query 8 CFSVFNF--KRTHCPRCFASGLCCHCSCCCCAPGL 40
C S+ NF + THC C A CHC C G+
Sbjct 553 CNSIINFLKENTHCEECHACAYRCHCDCLFSTAGV 587
> At1g12450
Length=303
Score = 31.6 bits (70), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 15/50 (30%), Positives = 30/50 (60%), Gaps = 2/50 (4%)
Query 385 VLFLRVSPFPNLVINAASPVLDVPFGAFFVATWLGLMPNSALFVSMGSAL 434
V +RVSPFP ++ N + V +G + + + +G++P +FVS+ + +
Sbjct 194 VTLIRVSPFPYIIYNYCALATGVHYGPYILGSLVGMVPE--IFVSIYTGI 241
> CE29511
Length=2104
Score = 31.6 bits (70), Expect = 4.5, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 306 PTESSSACGSGCSDISSSSNIASNDKSSSAAVQLKQKTQTSAE 348
PT +C S C D+SS N+ K S AA Q K+ S +
Sbjct 348 PTGYICSCNSNCIDVSSRYNLPPGRKCSVAANQCSDKSLNSCD 390
> At4g12000
Length=306
Score = 30.4 bits (67), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 22/88 (25%), Positives = 42/88 (47%), Gaps = 6/88 (6%)
Query 385 VLFLRVSPFPNLVINAASPVLDVPFGAFFVATWLGLMPNSALFVSMGSALGSL-DSLSSG 443
V +R+SPFP V N + V FG + + +G+ P + + G + +L D+ ++
Sbjct 200 VTLIRISPFPFAVYNYCAVATRVKFGPYMAGSLVGMAPEIFVAIYTGILIRTLADASTAE 259
Query 444 WRPLAVF-----VFGAFAVVLLKTALRK 466
+ L++ +FG A V+ + K
Sbjct 260 QKGLSILQIVLNIFGFVATVVTTVLITK 287
Lambda K H
0.327 0.138 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 12860921574
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40