bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2738_orf1
Length=162
Score E
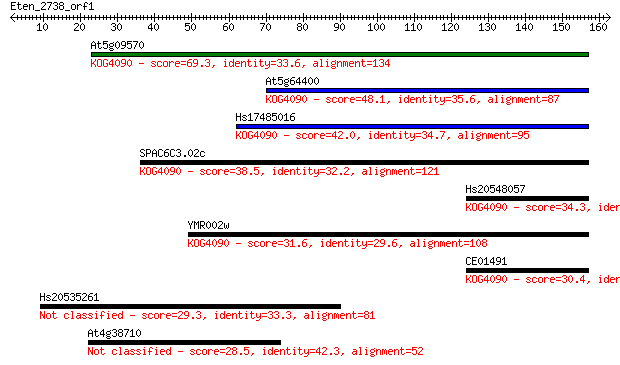
Sequences producing significant alignments: (Bits) Value
At5g09570 69.3 3e-12
At5g64400 48.1 6e-06
Hs17485016 42.0 5e-04
SPAC6C3.02c 38.5 0.005
Hs20548057 34.3 0.12
YMR002w 31.6 0.71
CE01491 30.4 1.5
Hs20535261 29.3 3.3
At4g38710 28.5 5.7
> At5g09570
Length=139
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 45/141 (31%), Positives = 64/141 (45%), Gaps = 20/141 (14%)
Query 23 RSSSGGGFFGSRSARPAAPAPVAAPP------SPPPVAGSSSGGGGFMANMVGNVASGMA 76
R SSGG S RP+ PA +PP +PPP S GG F+ N+ ++ G+A
Sbjct 3 RGSSGG----RSSYRPSRPAAARSPPPQSVNRAPPPATAQPSSGGSFLGNIGASITEGLA 58
Query 77 SGVGFGVAQRAVDAVMGPRSMEVVHTNTAPAAPAPQQPMQLGMQQQNPCAAYQEELNQCM 136
G G R VD+VMGPR+ + H P+ M C + + C+
Sbjct 59 WGTGTAFGHRVVDSVMGPRTFK--HETVVSQVPSAANTMTA-------CDIHSKAFQDCV 109
Query 137 TRH-TDITLCQNYLDNLKACQ 156
+DI+ CQ Y+D L C+
Sbjct 110 NHFGSDISKCQFYMDMLSECK 130
> At5g64400
Length=144
Score = 48.1 bits (113), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 31/88 (35%), Positives = 47/88 (53%), Gaps = 5/88 (5%)
Query 70 NVASGMASGVGFGVAQRAVDAVMGPRSMEVVHTNTAPAAPAPQQPMQLGMQQQNPCAAYQ 129
+A GMA G G VA RAVD+VMGPR+++ A A+ AP L + C +
Sbjct 52 TIAQGMAFGTGSAVAHRAVDSVMGPRTIQHEAVEAASASAAPAGSAML----SSTCDIHA 107
Query 130 EELNQCMTRH-TDITLCQNYLDNLKACQ 156
+ C+ + ++I+ CQ Y+D L C+
Sbjct 108 KAFQDCIGSYGSEISKCQFYMDMLSECR 135
> Hs17485016
Length=142
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 33/100 (33%), Positives = 46/100 (46%), Gaps = 14/100 (14%)
Query 62 GFMANMVGNVASGMASGVGFG-VAQRAVDAVMGPRSME----VVHTNTAPAAPAPQQPMQ 116
G MA M A+G+A G G V A+ S E V PAAP QP+Q
Sbjct 43 GLMAQMA-TTAAGVAVGSAVGHVMGSALTGAFSGGSSEPSQPAVQQAPTPAAP---QPLQ 98
Query 117 LGMQQQNPCAAYQEELNQCMTRHTDITLCQNYLDNLKACQ 156
+G PCA + C T +D++LC+ + + LK C+
Sbjct 99 MG-----PCAYEIRQFLDCSTTQSDLSLCEGFSEALKQCK 133
> SPAC6C3.02c
Length=172
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 39/137 (28%), Positives = 55/137 (40%), Gaps = 19/137 (13%)
Query 36 ARPAAPAPVAAPPSPPPVAGSSSGGGGFMANMVGNVAS-GMASGVGFGVAQRAVDAVMG- 93
A P AP+ V P P V G SS GF N+V A G+ S +G V G
Sbjct 32 APPPAPSRVQQAPPPTAVQGGSS--PGFFGNLVSTAAGVGIGSAIGHTVGSVITGGFSGS 89
Query 94 -----PRSMEV---VHTNTAPAA------PAPQQPMQLGMQQQNPCAAYQEELNQCMTRH 139
P V ++N+ P A P+ + + +N C + C+ H
Sbjct 90 GSNNAPADTSVPQSSYSNSVPEAAYGSAPPSTFASSAISEEAKNACKGDAKMFADCINEH 149
Query 140 TDITLCQNYLDNLKACQ 156
+ + C YL+ LKACQ
Sbjct 150 -EFSQCSYYLEQLKACQ 165
> Hs20548057
Length=278
Score = 34.3 bits (77), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 12/33 (36%), Positives = 18/33 (54%), Gaps = 0/33 (0%)
Query 124 PCAAYQEELNQCMTRHTDITLCQNYLDNLKACQ 156
PC E+ +C H DI LC+ + + LK C+
Sbjct 240 PCFYEIEQFLECAQNHGDIKLCEGFNEVLKQCR 272
> YMR002w
Length=156
Score = 31.6 bits (70), Expect = 0.71, Method: Compositional matrix adjust.
Identities = 32/114 (28%), Positives = 43/114 (37%), Gaps = 10/114 (8%)
Query 49 SPPPVAGSSSGGGGFMANMVGN---VASGMASGVGFGVAQRAVDAVMGPRS--MEVVHTN 103
S PP AG+ + G A M VA G G G + + G S +E N
Sbjct 41 SHPPAAGAQTRQPGMFAQMASTAAGVAVGSTIGHTLGAGITGMFSGSGSDSAPVEQQQQN 100
Query 104 TAPAAPAPQQPMQLGMQQQNPCAAYQEELNQCMTRHT-DITLCQNYLDNLKACQ 156
A + Q QLG C +C+ + + +C YL LKACQ
Sbjct 101 MANTSGQTQTDQQLG----RTCEIDARNFTRCLDENNGNFQICDYYLQQLKACQ 150
> CE01491
Length=154
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 9/33 (27%), Positives = 15/33 (45%), Gaps = 0/33 (0%)
Query 124 PCAAYQEELNQCMTRHTDITLCQNYLDNLKACQ 156
PC + C +D++LC + D K C+
Sbjct 118 PCEFEWRQFVDCAQNQSDVSLCNGFNDIFKQCK 150
> Hs20535261
Length=536
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 27/91 (29%), Positives = 36/91 (39%), Gaps = 11/91 (12%)
Query 9 VLKMPRQRSSSGARRSSSGGGFFGSRSARPAAPAPVAAPPSP----------PPVAGSSS 58
LKM + R+ +GGG +R ARP AP + P P P + S
Sbjct 43 TLKMCTDKLKISGLRAHAGGGATWAR-ARPTAPGRITRPGLPWQQRRRAARRPAMRESQD 101
Query 59 GGGGFMANMVGNVASGMASGVGFGVAQRAVD 89
G N VG+ A+ +G FGV D
Sbjct 102 AAGAHGWNRVGSTATKWFTGAPFGVQSHRFD 132
> At4g38710
Length=452
Score = 28.5 bits (62), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 22/54 (40%), Positives = 26/54 (48%), Gaps = 2/54 (3%)
Query 22 RRSSSGGGFFGSRSARPAAPAP--VAAPPSPPPVAGSSSGGGGFMANMVGNVAS 73
R SGGGFF S+S A V+ PS P SS+GGGG G+ S
Sbjct 184 RERGSGGGFFESQSQSKADEVDSWVSTKPSEPRRFVSSNGGGGDRFEKRGSFES 237
Lambda K H
0.317 0.129 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2244926132
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40