bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2734_orf1
Length=141
Score E
Sequences producing significant alignments: (Bits) Value
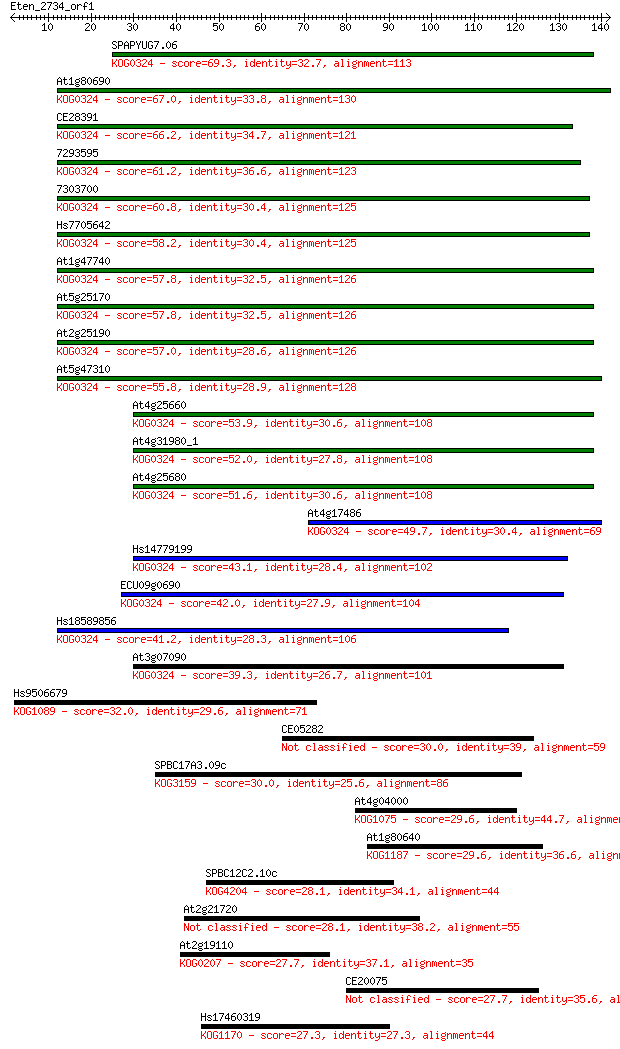
SPAPYUG7.06 69.3 2e-12
At1g80690 67.0 1e-11
CE28391 66.2 2e-11
7293595 61.2 6e-10
7303700 60.8 8e-10
Hs7705642 58.2 5e-09
At1g47740 57.8 7e-09
At5g25170 57.8 7e-09
At2g25190 57.0 1e-08
At5g47310 55.8 2e-08
At4g25660 53.9 9e-08
At4g31980_1 52.0 4e-07
At4g25680 51.6 4e-07
At4g17486 49.7 2e-06
Hs14779199 43.1 2e-04
ECU09g0690 42.0 4e-04
Hs18589856 41.2 7e-04
At3g07090 39.3 0.002
Hs9506679 32.0 0.36
CE05282 30.0 1.5
SPBC17A3.09c 30.0 1.7
At4g04000 29.6 2.1
At1g80640 29.6 2.1
SPBC12C2.10c 28.1 5.3
At2g21720 28.1 5.9
At2g19110 27.7 8.0
CE20075 27.7 8.1
Hs17460319 27.3 8.4
> SPAPYUG7.06
Length=191
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/118 (31%), Positives = 59/118 (50%), Gaps = 11/118 (9%)
Query 25 WCSPCGLYHTSVQIGELEYSFGE-----DSGVMCSEHNPRIDGIHSFLDGTYEYSLHMGA 79
W G+YHT + + EY+FG +GV + P ++G + S+ +
Sbjct 11 WTLGLGIYHTGLVLEGKEYAFGAHEIPGSTGVFATMPRPPLEGCR------WRCSIALPN 64
Query 80 CNLSVPELHKVISSLQKSFPGSSYDLIRNNCNHFSDALLKSIVGRGIPPHINRASRYG 137
C L P++ +++ L + F G SY L+ NCNHF++A + G IP +NR SR G
Sbjct 65 CTLPKPDVDRILIRLSQEFTGLSYSLLERNCNHFTNAAAIELTGSPIPSFLNRISRIG 122
> At1g80690
Length=231
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 44/138 (31%), Positives = 72/138 (52%), Gaps = 17/138 (12%)
Query 12 VFLNIYK---IHGSPDWCSPCGLYHTSVQIGELEYSFGE----DSGVMCSEHNPR-IDGI 63
V+LN+Y I+G W G+YH+ V++ +EY++G +G+ E P+ +G
Sbjct 17 VYLNVYDLTPINGYAYWLG-LGVYHSGVEVHGIEYAYGAHEYPSTGIF--EGEPKQCEGF 73
Query 64 HSFLDGTYEYSLHMGACNLSVPELHKVISSLQKSFPGSSYDLIRNNCNHFSDALLKSIVG 123
T+ S+ +G +L E+ + L ++ GSSY+LI NCNHF D + G
Sbjct 74 ------TFRKSILIGKTDLGPLEVRATMEQLADNYKGSSYNLITKNCNHFCDETCIKLTG 127
Query 124 RGIPPHINRASRYGQWLG 141
IP +NR +R G++ G
Sbjct 128 NPIPSWVNRLARIGKFSG 145
> CE28391
Length=334
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 42/127 (33%), Positives = 70/127 (55%), Gaps = 9/127 (7%)
Query 12 VFLNIYKIHGSPDWCSP--CGLYHTSVQIGELEYSFG----EDSGVMCSEHNPRIDGIHS 65
V LN+Y ++ D+ S G++H+ +++ +EY++G + SGV E++P+ D
Sbjct 32 VRLNVYDMYWLNDYASNIGVGIFHSGIEVFGVEYAYGGHPYQFSGVF--ENSPQ-DAEEL 88
Query 66 FLDGTYEYSLHMGACNLSVPELHKVISSLQKSFPGSSYDLIRNNCNHFSDALLKSIVGRG 125
++ S+ +G S ++ K+I SL + F G Y LI NCNHFS L + + G+
Sbjct 89 GETFKFKESIVVGETERSTSDIRKLIKSLGEDFRGDRYHLISRNCNHFSAVLARELTGKD 148
Query 126 IPPHINR 132
IP INR
Sbjct 149 IPGWINR 155
> 7293595
Length=183
Score = 61.2 bits (147), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 45/135 (33%), Positives = 64/135 (47%), Gaps = 20/135 (14%)
Query 12 VFLNIYKIHGSPDWCSPCGL--YHTSVQIGELEYSFGEDSGVMCSEHNPRIDGIHSF--L 67
V LNIY + S ++ P GL +H+ VQ+ EY+F N I GI
Sbjct 29 VMLNIYDLSTSNNYTFPLGLGVFHSGVQLYGREYAF--------LALNLSISGIFEIHPC 80
Query 68 DGTYEYSLH--------MGACNLSVPELHKVISSLQKSFPGSSYDLIRNNCNHFSDALLK 119
+G E H +G + + E+ +VI+ L F G+SY L NCNHFS+ L
Sbjct 81 NGQEELGEHFRFRKSILLGYTDFTCAEVKRVINLLGFEFRGTSYHLTSKNCNHFSNCLAH 140
Query 120 SIVGRGIPPHINRAS 134
+ GR IP +NR +
Sbjct 141 LVCGRKIPRWVNRLA 155
> 7303700
Length=205
Score = 60.8 bits (146), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 38/133 (28%), Positives = 71/133 (53%), Gaps = 13/133 (9%)
Query 12 VFLNIYKIHGSPDWCSPCGL--YHTSVQIGELEYSFGED----SGVMCSEHNPRIDGIHS 65
V LN+Y ++ ++ + GL +H+ V+ E+++G +GV E +PR H
Sbjct 39 VILNVYDMYWINEYTTSIGLGVFHSGVEAFGTEFAYGGHPFPFTGVF--EISPRD---HD 93
Query 66 FLDGTYEY--SLHMGACNLSVPELHKVISSLQKSFPGSSYDLIRNNCNHFSDALLKSIVG 123
L +++ S+ +G + + E+ +++ L F G Y L+ NNCNHFS +L + + G
Sbjct 94 ELGDQFQFRQSIQIGCTDFTYEEVRRIVEELGNQFRGDRYHLMNNNCNHFSGSLTQILCG 153
Query 124 RGIPPHINRASRY 136
+ IP +NR + +
Sbjct 154 QEIPSWVNRLAHF 166
> Hs7705642
Length=193
Score = 58.2 bits (139), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 38/133 (28%), Positives = 75/133 (56%), Gaps = 13/133 (9%)
Query 12 VFLNIYKIHGSPDWCSPCGL--YHTSVQIGELEYSFGED----SGVMCSEHNPRIDGIHS 65
V LN+Y ++ ++ S G+ +H+ +++ E+++G SG+ E +P G S
Sbjct 7 VVLNVYDMYWMNEYTSSIGIGVFHSGIEVYGREFAYGGHPYPFSGIF--EISP---GNAS 61
Query 66 FLDGTYEY--SLHMGACNLSVPELHKVISSLQKSFPGSSYDLIRNNCNHFSDALLKSIVG 123
L T+++ ++ +G+ + ++ K++ L K + G++Y L+ NCNHFS AL + + G
Sbjct 62 ELGETFKFKEAVVLGSTDFLEDDIEKIVEELGKEYKGNAYHLMHKNCNHFSSALSEILCG 121
Query 124 RGIPPHINRASRY 136
+ IP INR + +
Sbjct 122 KEIPRWINRLAYF 134
> At1g47740
Length=279
Score = 57.8 bits (138), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 41/133 (30%), Positives = 68/133 (51%), Gaps = 15/133 (11%)
Query 12 VFLNIYK---IHGSPDWCSPCGLYHTSVQIGELEYSFGE----DSGVMCSEHNPRIDGIH 64
V+LN+Y I+G W G++H+ V++ +EY+FG SGV E PR
Sbjct 71 VYLNVYDLTPINGYIYWAG-LGIFHSGVEVHGVEYAFGAHDYATSGVF--EVEPRQCPGF 127
Query 65 SFLDGTYEYSLHMGACNLSVPELHKVISSLQKSFPGSSYDLIRNNCNHFSDALLKSIVGR 124
F + S+ +G NL+ ++ + + + S+ G+ Y LI NCNHF + + G+
Sbjct 128 KF-----KKSIFIGTTNLNPTQVREFMEDMACSYYGNMYHLIVKNCNHFCQDVCYKLTGK 182
Query 125 GIPPHINRASRYG 137
IP +NR ++ G
Sbjct 183 KIPKWVNRLAQIG 195
> At5g25170
Length=211
Score = 57.8 bits (138), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 41/134 (30%), Positives = 63/134 (47%), Gaps = 20/134 (14%)
Query 12 VFLNIYK---IHGSPDWCSPCGLYHTSVQIGELEYSFGEDSGVMCSEHNPRIDGIHSFLD 68
V+LN+Y I+G W G+YH+ V E+EY FG H+ GI
Sbjct 16 VYLNVYDLTPINGYAYWLG-LGIYHSGV---EVEYGFGA--------HDHSTTGIFEVEP 63
Query 69 G-----TYEYSLHMGACNLSVPELHKVISSLQKSFPGSSYDLIRNNCNHFSDALLKSIVG 123
T+ S+ +G +L + + L + + G+SY LI NCNHF + + +
Sbjct 64 KQCPGFTFRKSILIGRTDLDPENVRVFMEKLAEEYSGNSYHLITKNCNHFCNDVCVQLTR 123
Query 124 RGIPPHINRASRYG 137
R IP +NR +R+G
Sbjct 124 RSIPSWVNRLARFG 137
> At2g25190
Length=240
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 36/130 (27%), Positives = 64/130 (49%), Gaps = 9/130 (6%)
Query 12 VFLNIYKIH--GSPDWCSPCGLYHTSVQIGELEYSFG--EDSGVMCSEHNPRIDGIHSFL 67
V+LN+Y + + + G++H+ V++ +EY+FG E S E P+
Sbjct 19 VYLNVYDLTPMNAYGYWLGLGVFHSGVEVHGVEYAFGAHESSSTGIFEVEPK-----KCP 73
Query 68 DGTYEYSLHMGACNLSVPELHKVISSLQKSFPGSSYDLIRNNCNHFSDALLKSIVGRGIP 127
T+ S+ +G +L E+ + L + + G+ Y LI NCNHF + + + + IP
Sbjct 74 GFTFRKSILVGKTDLVAKEVRVFMEKLAEEYQGNKYHLITRNCNHFCNEVCLKLAQKSIP 133
Query 128 PHINRASRYG 137
+NR +R G
Sbjct 134 RWVNRLARLG 143
> At5g47310
Length=245
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 37/134 (27%), Positives = 65/134 (48%), Gaps = 13/134 (9%)
Query 12 VFLNIYKIHGSPDWCS--PCGLYHTSVQIGELEYSFG----EDSGVMCSEHNPRIDGIHS 65
V+LN+Y + ++ G++H+ ++ EY +G SGV E PR S
Sbjct 30 VYLNVYDLTPVNNYLYWFGLGIFHSGIEAHGFEYGYGAHEYSSSGVF--EVEPR-----S 82
Query 66 FLDGTYEYSLHMGACNLSVPELHKVISSLQKSFPGSSYDLIRNNCNHFSDALLKSIVGRG 125
+ S+ +G ++S + + L + + G +Y LI NCNHF++ + + G+
Sbjct 83 CPGFIFRRSVLLGTTSMSRSDFRSFMEKLSRKYHGDTYHLIAKNCNHFTEEVCLQVTGKP 142
Query 126 IPPHINRASRYGQW 139
IP INR +R G +
Sbjct 143 IPGWINRMARVGSF 156
> At4g25660
Length=255
Score = 53.9 bits (128), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 33/114 (28%), Positives = 60/114 (52%), Gaps = 15/114 (13%)
Query 30 GLYHTSVQI-GELEYSFG---EDSGVMC--SEHNPRIDGIHSFLDGTYEYSLHMGACNLS 83
G++H+++Q+ G E+S+G + +GV S NP TY + +G + +
Sbjct 38 GIFHSAIQVYGNDEWSYGYCEQGTGVFSCPSGKNPMY---------TYREKIVLGKTDCT 88
Query 84 VPELHKVISSLQKSFPGSSYDLIRNNCNHFSDALLKSIVGRGIPPHINRASRYG 137
+ +++++ L + +PG +YDL+ NCNHF D L + IP +NR + G
Sbjct 89 IFMVNQILRELSREWPGHTYDLLSKNCNHFCDVLCDRLGVPKIPGWVNRFAHAG 142
> At4g31980_1
Length=266
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 30/110 (27%), Positives = 53/110 (48%), Gaps = 7/110 (6%)
Query 30 GLYHTSVQIGELEYSFG--EDSGVMCSEHNPRIDGIHSFLDGTYEYSLHMGACNLSVPEL 87
G+YH+ +++ +EY +G E S E P+ T+ S+ +G + E+
Sbjct 39 GIYHSGLEVHGVEYGYGAHEKSSSGIFEVEPK-----KCPGFTFRKSILVGETEMKAKEV 93
Query 88 HKVISSLQKSFPGSSYDLIRNNCNHFSDALLKSIVGRGIPPHINRASRYG 137
+ L + + G+ Y LI NCNHF + + + + IP +NR +R G
Sbjct 94 RSFMEKLSEEYQGNKYHLITRNCNHFCNHVSLKLTHKSIPSWVNRLARLG 143
> At4g25680
Length=252
Score = 51.6 bits (122), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 33/114 (28%), Positives = 59/114 (51%), Gaps = 15/114 (13%)
Query 30 GLYHTSVQI-GELEYSFG---EDSGVMC--SEHNPRIDGIHSFLDGTYEYSLHMGACNLS 83
G++H+++Q+ G E+S+G +GV S NP TY + +G + +
Sbjct 38 GIFHSAIQVYGNDEWSYGYCELGTGVFSCPSGKNPMY---------TYREKIVLGKTDCT 88
Query 84 VPELHKVISSLQKSFPGSSYDLIRNNCNHFSDALLKSIVGRGIPPHINRASRYG 137
+ +++++ L + +PG +YDL+ NCNHF D L + IP +NR + G
Sbjct 89 IFMVNQMLRELSREWPGHTYDLLSKNCNHFCDVLCDRLGVPKIPGWVNRFANAG 142
> At4g17486
Length=246
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 38/69 (55%), Gaps = 0/69 (0%)
Query 71 YEYSLHMGACNLSVPELHKVISSLQKSFPGSSYDLIRNNCNHFSDALLKSIVGRGIPPHI 130
+ S+ +G ++S + + L + + G +Y LI NCNHF++ + + G+ IP I
Sbjct 108 FRRSVLLGTTSMSRSDFRSYMEKLSRKYHGDTYHLIAKNCNHFTEEVCLQLTGKPIPGWI 167
Query 131 NRASRYGQW 139
NR +R G +
Sbjct 168 NRLARVGSF 176
> Hs14779199
Length=168
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 52/103 (50%), Gaps = 9/103 (8%)
Query 30 GLYHTSVQIGELEYSFGEDSGVMCSEHNPRIDGIHSFLDGTYEYSLHMGACNLSVPELHK 89
G++HTS+ + + E+ FG C + S +D +G+ ++ +
Sbjct 35 GIWHTSIVVHKDEFFFGSGGISSCPPGGTLLGPPDSVVD--------VGSTEVTEEIFLE 86
Query 90 VISSLQKS-FPGSSYDLIRNNCNHFSDALLKSIVGRGIPPHIN 131
+SSL +S F G +Y+L +NCN FS+ + + + GR IP +I
Sbjct 87 YLSSLGESLFRGEAYNLFEHNCNTFSNEVAQFLTGRKIPSYIT 129
> ECU09g0690
Length=150
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 29/108 (26%), Positives = 51/108 (47%), Gaps = 17/108 (15%)
Query 27 SPCGLYHTSVQIGELEYSFGEDSGVMCSEHNPRIDG----IHSFLDGTYEYSLHMGACNL 82
S ++HTS+++ EY F +G+M + I G IH +GA ++
Sbjct 29 SEACIWHTSIEVYGTEYFF--QNGIMKARPGSTIYGTPLKIHD-----------LGATDI 75
Query 83 SVPELHKVISSLQKSFPGSSYDLIRNNCNHFSDALLKSIVGRGIPPHI 130
+ S+ + F Y L++NNCN+F++ L +V + IP +I
Sbjct 76 PEVVFEDFLFSIAEDFAPHKYHLLKNNCNNFTNTLALYLVEKSIPEYI 123
> Hs18589856
Length=154
Score = 41.2 bits (95), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 30/113 (26%), Positives = 60/113 (53%), Gaps = 12/113 (10%)
Query 12 VFLNIYKIHGSPDWCSPCGL--YHTSVQIGELEYSFGED----SGVMCSEHNPRIDGIHS 65
V LN+ ++ ++ S G+ +H+ +++ E+++G SG+ E +P G S
Sbjct 7 VVLNVCDMYWMKEYTSSIGIGVFHSEIEVYGREFAYGGHPYPFSGIF--EISP---GNAS 61
Query 66 FLDGTYEYS-LHMGACNLSVPELHKVISSLQKSFPGSSYDLIRNNCNHFSDAL 117
L +++ + +G+ + ++ K++ L K + G+ Y L+ NCNHFS AL
Sbjct 62 ELGEPFKFKEVVLGSTDFLEDDIEKIVEELGKEYKGNVYHLMHKNCNHFSSAL 114
> At3g07090
Length=265
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/101 (26%), Positives = 52/101 (51%), Gaps = 9/101 (8%)
Query 30 GLYHTSVQIGELEYSFGEDSGVMCSEHNPRIDGIHSFLDGTYEYSLHMGACNLSVPELHK 89
G++HT + + EY FG G+ +H P + GT ++ +G ++
Sbjct 34 GVWHTGIVVYGNEYFFG--GGI---QHLP----VGRTPYGTPIRTIELGLSHVPKDVFEM 84
Query 90 VISSLQKSFPGSSYDLIRNNCNHFSDALLKSIVGRGIPPHI 130
+ + + SY+L+ +NCN+FS+ + + +VG+GIP +I
Sbjct 85 YLEEISPRYTAESYNLLTHNCNNFSNEVAQFLVGKGIPDYI 125
> Hs9506679
Length=637
Score = 32.0 bits (71), Expect = 0.36, Method: Composition-based stats.
Identities = 21/71 (29%), Positives = 32/71 (45%), Gaps = 10/71 (14%)
Query 2 YFVNRMPGPEVFLNIYKIHGSPDWCSPCGLYHTSVQIGELEYSFGEDSGVMCSEHNPRID 61
YFV P PE + ++ HG P WC C +++ L +D G++ +
Sbjct 242 YFVVPTPLPEENVQRFQGHGIPIWCWSCHNGSALLKMSALPKE--QDDGILQIQ------ 293
Query 62 GIHSFLDGTYE 72
SFLDG Y+
Sbjct 294 --KSFLDGIYK 302
> CE05282
Length=677
Score = 30.0 bits (66), Expect = 1.5, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 39/76 (51%), Gaps = 17/76 (22%)
Query 65 SFLDGTYEYSL-HMG--ACNLSVPELHKVISSLQK----SFPGSSYDLIRNN-------- 109
SF+D E+SL H+ + +V ELHK++ SLQ F G+ D++R N
Sbjct 273 SFVDTATEFSLEHLEDLIADGAVDELHKLLKSLQTWPEWMFTGNRQDIVRINDVLRFIVE 332
Query 110 --CNHFSDALLKSIVG 123
C+ S+ ++++ G
Sbjct 333 NLCSEESEIIIRNFCG 348
> SPBC17A3.09c
Length=363
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 22/86 (25%), Positives = 37/86 (43%), Gaps = 18/86 (20%)
Query 35 SVQIGELEYSFGEDSGVMCSEHNPRIDGIHSFLDGTYEYSLHMGACNLSVPELHKVISSL 94
SV + E+S E++ +M R G+H+ L+ ++ V++
Sbjct 108 SVLMNREEFSHTENASIMI--QALRNLGVHARLNQRHDI----------------VLAQS 149
Query 95 QKSFPGSSYDLIRNNCNHFSDALLKS 120
Q+ GS+Y + RN C H LL S
Sbjct 150 QRKISGSAYKISRNRCYHHGTMLLNS 175
> At4g04000
Length=1077
Score = 29.6 bits (65), Expect = 2.1, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 21/41 (51%), Gaps = 3/41 (7%)
Query 82 LSVPELHKV---ISSLQKSFPGSSYDLIRNNCNHFSDALLK 119
L V ELH + I SL SF ++ I NH +DAL K
Sbjct 1028 LQVKELHGILHDILSLSSSFDACTFTFINREFNHQADALAK 1068
> At1g80640
Length=444
Score = 29.6 bits (65), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 85 PELHKVISSLQKSFPGSSYDLIRNNCNHFSDALLKSIVGRG 125
P +HK+ S + + P Y L+ + N FSD+ + S GRG
Sbjct 121 PTVHKIDSVRKGTIPVYEYQLLESATNKFSDSNVLSRGGRG 161
> SPBC12C2.10c
Length=781
Score = 28.1 bits (61), Expect = 5.3, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 23/44 (52%), Gaps = 1/44 (2%)
Query 47 EDSGVMCSEHNPRIDGIHSFLDGTYEYSLHMGACNLSVPELHKV 90
EDSG + N + +H + YEY H+GA N+ EL ++
Sbjct 636 EDSGFIAHRKNQFEENLHKLEEERYEYDRHIGA-NMRFIELLQI 678
> At2g21720
Length=703
Score = 28.1 bits (61), Expect = 5.9, Method: Composition-based stats.
Identities = 21/61 (34%), Positives = 30/61 (49%), Gaps = 6/61 (9%)
Query 42 EYSFGEDSGVMCSEHNPR----IDGIHSF--LDGTYEYSLHMGACNLSVPELHKVISSLQ 95
E S GED+ V P I+G ++F L + LH A ++ V E+HK + LQ
Sbjct 196 EPSVGEDAFVYLGSIIPLPVDIINGRYTFETLTAPTGHQLHFPAYDMFVKEIHKCMKHLQ 255
Query 96 K 96
K
Sbjct 256 K 256
> At2g19110
Length=1172
Score = 27.7 bits (60), Expect = 8.0, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 41 LEYSFGEDSGVMCSEHNPRIDGIHSFLDGTYEYSL 75
L+ S+ + G+ C+ P I+ I LDG EYS+
Sbjct 16 LQKSYFDVLGICCTSEVPIIENILKSLDGVKEYSV 50
> CE20075
Length=434
Score = 27.7 bits (60), Expect = 8.1, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 21/45 (46%), Gaps = 1/45 (2%)
Query 80 CNLSVPELHKVISSLQKSFPGSSYDLIRNNCNHFSDALLKSIVGR 124
CN ELH ++ SL + S L + N S KS+VGR
Sbjct 176 CNAKPHELHPILQSL-ATLQAGSLHLTEDEINSISGYDFKSMVGR 219
> Hs17460319
Length=1079
Score = 27.3 bits (59), Expect = 8.4, Method: Composition-based stats.
Identities = 12/44 (27%), Positives = 21/44 (47%), Gaps = 0/44 (0%)
Query 46 GEDSGVMCSEHNPRIDGIHSFLDGTYEYSLHMGACNLSVPELHK 89
GE + +E+NP + G+H + + H C S+P L +
Sbjct 106 GEIYKIPAAENNPFLVGMHCWYSSYSHRTQHCNVCRESIPALSR 149
Lambda K H
0.321 0.141 0.459
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1608078824
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40