bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2724_orf1
Length=123
Score E
Sequences producing significant alignments: (Bits) Value
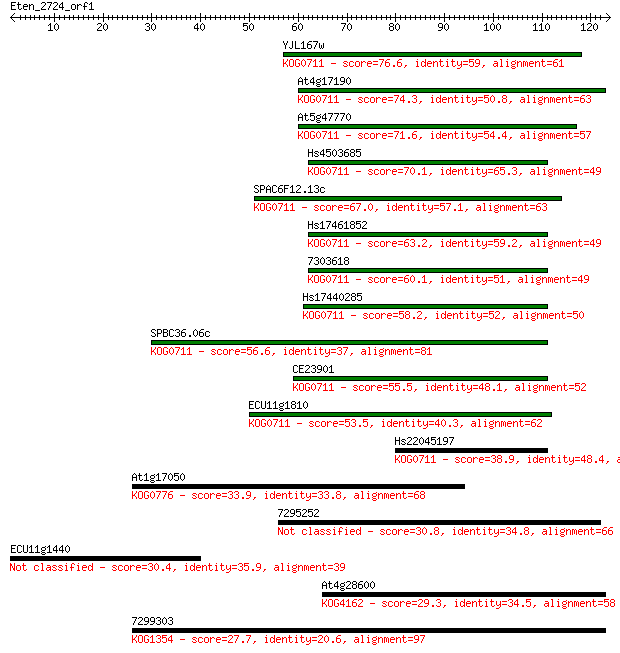
YJL167w 76.6 9e-15
At4g17190 74.3 5e-14
At5g47770 71.6 3e-13
Hs4503685 70.1 1e-12
SPAC6F12.13c 67.0 8e-12
Hs17461852 63.2 1e-10
7303618 60.1 1e-09
Hs17440285 58.2 4e-09
SPBC36.06c 56.6 1e-08
CE23901 55.5 3e-08
ECU11g1810 53.5 9e-08
Hs22045197 38.9 0.002
At1g17050 33.9 0.071
7295252 30.8 0.64
ECU11g1440 30.4 0.82
At4g28600 29.3 2.1
7299303 27.7 4.9
> YJL167w
Length=352
Score = 76.6 bits (187), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 36/64 (56%), Positives = 45/64 (70%), Gaps = 4/64 (6%)
Query 57 SELCVLGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFL---IYS 113
++ +LGW +ELLQA FLVADD MD + TRRG+ CWY+ PEVG AINDA L IY
Sbjct 80 EKVAILGWCIELLQAYFLVADDMMDKSITRRGQPCWYKVPEVGEI-AINDAFMLEAAIYK 138
Query 114 VHQS 117
+ +S
Sbjct 139 LLKS 142
> At4g17190
Length=342
Score = 74.3 bits (181), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 32/63 (50%), Positives = 41/63 (65%), Gaps = 1/63 (1%)
Query 60 CVLGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFLIYSVHQSDP 119
C LGW +E LQA FLV DD MD + TRRG+ CW+R+P+VG AIND + L +H+
Sbjct 76 CALGWCIEWLQAYFLVLDDIMDNSVTRRGQPCWFRKPKVGMI-AINDGILLRNHIHRILK 134
Query 120 PSF 122
F
Sbjct 135 KHF 137
> At5g47770
Length=384
Score = 71.6 bits (174), Expect = 3e-13, Method: Composition-based stats.
Identities = 31/57 (54%), Positives = 39/57 (68%), Gaps = 1/57 (1%)
Query 60 CVLGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFLIYSVHQ 116
C LGW +E LQA FLV DD MD + TRRG+ CW+R P+VG AIND + L +H+
Sbjct 118 CALGWCIEWLQAYFLVLDDIMDNSVTRRGQPCWFRVPQVGMV-AINDGILLRNHIHR 173
> Hs4503685
Length=419
Score = 70.1 bits (170), Expect = 1e-12, Method: Composition-based stats.
Identities = 32/49 (65%), Positives = 38/49 (77%), Gaps = 1/49 (2%)
Query 62 LGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFL 110
+GW VELLQA FLVADD MD + TRRG+ CWY++P VG +AINDA L
Sbjct 154 VGWCVELLQAFFLVADDIMDSSLTRRGQICWYQKPGVG-LDAINDANLL 201
> SPAC6F12.13c
Length=347
Score = 67.0 bits (162), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 36/63 (57%), Positives = 44/63 (69%), Gaps = 2/63 (3%)
Query 51 LDKAFGSELCVLGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFL 110
LD+A + VLGW VELLQ+ FL+ADD MD + TRRG+ CWY P VG AINDA F+
Sbjct 69 LDEAAYMKAAVLGWMVELLQSFFLIADDIMDASKTRRGQPCWYLMPGVGNI-AINDA-FM 126
Query 111 IYS 113
+ S
Sbjct 127 VES 129
> Hs17461852
Length=266
Score = 63.2 bits (152), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 29/49 (59%), Positives = 38/49 (77%), Gaps = 1/49 (2%)
Query 62 LGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFL 110
+GW VELLQA FLVADD MD + +RRG+ WY++P +G +AINDA+ L
Sbjct 88 VGWCVELLQAFFLVADDIMDSSVSRRGQIWWYQKPGMG-LDAINDAMLL 135
> 7303618
Length=419
Score = 60.1 bits (144), Expect = 1e-09, Method: Composition-based stats.
Identities = 25/49 (51%), Positives = 36/49 (73%), Gaps = 1/49 (2%)
Query 62 LGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFL 110
LGW VE+LQ+ F+++DD MD + TRRG+ CW++ VG AINDA+ +
Sbjct 157 LGWCVEMLQSFFIISDDVMDNSTTRRGQPCWHKVENVG-LTAINDALMI 204
> Hs17440285
Length=258
Score = 58.2 bits (139), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 26/50 (52%), Positives = 35/50 (70%), Gaps = 1/50 (2%)
Query 61 VLGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFL 110
++GW VELLQA FLV DD D + T G+ CWY++ VG +A+NDA+ L
Sbjct 84 MVGWCVELLQAFFLVVDDITDSSLTHWGQICWYQKLGVG-LDAVNDAMLL 132
> SPBC36.06c
Length=351
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 30/81 (37%), Positives = 42/81 (51%), Gaps = 2/81 (2%)
Query 30 GGRNPAAPGSADDSFAAGEAALDKAFGSELCVLGWAVELLQAAFLVADDQMDGAFTRRGK 89
GG+N + L++A + +LGW +E+LQ FL+ADD MD + RRG
Sbjct 53 GGKNNRGLAVLQSLTSLINRELEEAEFRDAALLGWLIEILQGCFLMADDIMDQSIKRRGL 112
Query 90 ACWYRRPEVGPANAINDAVFL 110
CWY VG AIN++ L
Sbjct 113 DCWY--LVVGVRRAINESQLL 131
> CE23901
Length=352
Score = 55.5 bits (132), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 25/52 (48%), Positives = 36/52 (69%), Gaps = 1/52 (1%)
Query 59 LCVLGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFL 110
+C +E++Q+ +L+ADD MD + TRRGK CW+RR VG +AINDA +
Sbjct 81 VCEAAATLEIIQSFYLIADDIMDNSETRRGKPCWFRREGVG-MSAINDAFIM 131
> ECU11g1810
Length=292
Score = 53.5 bits (127), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 36/62 (58%), Gaps = 3/62 (4%)
Query 50 ALDKAFGSELCVLGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVF 109
AL+ E ++G++VELLQA L+ DD MD + RRGK C+Y + + DA F
Sbjct 47 ALNGEVSMENLIMGYSVELLQAVLLIVDDIMDNSKIRRGKPCYYLKR---GMKTVKDAFF 103
Query 110 LI 111
L+
Sbjct 104 LL 105
> Hs22045197
Length=184
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 22/31 (70%), Gaps = 1/31 (3%)
Query 80 MDGAFTRRGKACWYRRPEVGPANAINDAVFL 110
MD +FT RG+ CWY++P + +AIND + L
Sbjct 1 MDSSFTCRGQICWYQKPGMD-LDAINDVILL 30
> At1g17050
Length=336
Score = 33.9 bits (76), Expect = 0.071, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 36/85 (42%), Gaps = 17/85 (20%)
Query 26 VTISGGRNPAAPGSADDSFAAG-----------------EAALDKAFGSELCVLGWAVEL 68
++I G NP +A+ F+AG E A K E LG +E+
Sbjct 33 LSIVGAENPVLISAAEQIFSAGGKRMRPGLVFLVSRATAELAGLKELTVEHRRLGEIIEM 92
Query 69 LQAAFLVADDQMDGAFTRRGKACWY 93
+ A L+ DD +D + RRG+ +
Sbjct 93 IHTASLIHDDVLDESDMRRGRETVH 117
> 7295252
Length=338
Score = 30.8 bits (68), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 33/68 (48%), Gaps = 2/68 (2%)
Query 56 GSELCVLGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPA-NAINDAVFL-IYS 113
G +L +G V++L + L+ DD D + RRG + V NA N A+FL +
Sbjct 55 GEKLAQIGDIVQMLHNSSLLIDDIEDNSILRRGVPVAHSIYGVASTINAANYALFLALEK 114
Query 114 VHQSDPPS 121
V Q D P
Sbjct 115 VQQLDHPE 122
> ECU11g1440
Length=1303
Score = 30.4 bits (67), Expect = 0.82, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 1 STSAAAAAAPAFSQTGATCGTVARPVTISGGRNPAAPGS 39
+T+ ++ PA +Q A V P+ S GRNP PG+
Sbjct 1207 TTTPSSGRRPASNQAVAPANVVPGPMVPSSGRNPLKPGA 1245
> At4g28600
Length=710
Score = 29.3 bits (64), Expect = 2.1, Method: Composition-based stats.
Identities = 20/60 (33%), Positives = 27/60 (45%), Gaps = 2/60 (3%)
Query 65 AVELLQAAFLVADDQMDGAFTRRGKAC--WYRRPEVGPANAINDAVFLIYSVHQSDPPSF 122
AVELL + +AD D + R W PE AVFL+YS ++ PP+
Sbjct 184 AVELLPELWKLADSPRDAILSYRRALLNHWKLDPETTARIQKEYAVFLLYSGEEAVPPNL 243
> 7299303
Length=499
Score = 27.7 bits (60), Expect = 4.9, Method: Composition-based stats.
Identities = 20/97 (20%), Positives = 37/97 (38%), Gaps = 8/97 (8%)
Query 26 VTISGGRNPAAPGSADDSFAAGEAALDKAFGSELCVLGWAVELLQAAFLVADDQMDGAFT 85
+ I+G + PA+ G A F+ + ALD +++ D ++
Sbjct 49 ININGAKKPASNGEASWCFSQIKGALDDDVTD--------ADIISCVEFNHDGELLATGD 100
Query 86 RRGKACWYRRPEVGPANAINDAVFLIYSVHQSDPPSF 122
+ G+ ++R A + +YS QS P F
Sbjct 101 KGGRVVIFQRDPASKAANPRRGEYNVYSTFQSHEPEF 137
Lambda K H
0.318 0.130 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1191192512
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40