bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2721_orf2
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
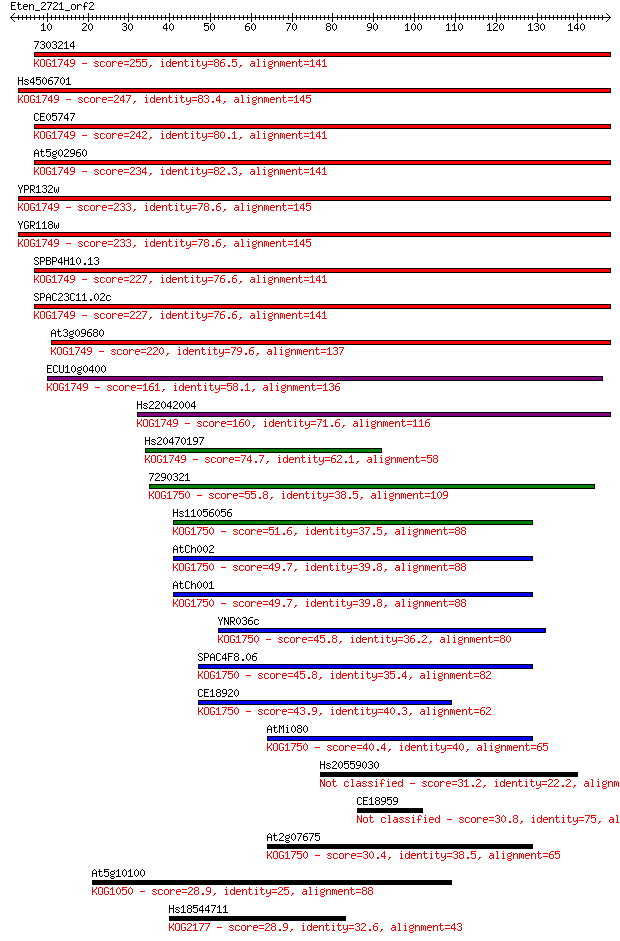
7303214 255 2e-68
Hs4506701 247 7e-66
CE05747 242 2e-64
At5g02960 234 4e-62
YPR132w 233 9e-62
YGR118w 233 9e-62
SPBP4H10.13 227 6e-60
SPAC23C11.02c 227 6e-60
At3g09680 220 7e-58
ECU10g0400 161 4e-40
Hs22042004 160 8e-40
Hs20470197 74.7 6e-14
7290321 55.8 3e-08
Hs11056056 51.6 5e-07
AtCh002 49.7 2e-06
AtCh001 49.7 2e-06
YNR036c 45.8 3e-05
SPAC4F8.06 45.8 3e-05
CE18920 43.9 1e-04
AtMi080 40.4 0.001
Hs20559030 31.2 0.73
CE18959 30.8 0.99
At2g07675 30.4 1.2
At5g10100 28.9 3.3
Hs18544711 28.9 4.1
> 7303214
Length=216
Score = 255 bits (652), Expect = 2e-68, Method: Compositional matrix adjust.
Identities = 122/141 (86%), Positives = 133/141 (94%), Gaps = 0/141 (0%)
Query 7 KPRGMRTARKLRNRRRVQKWADKQYKKAHLGTRWKANPFGGSSHAKGIVVEKLGIEAKQP 66
KPRG+RTARK N RR Q+WADK YKKAHLGTRWKANPFGG+SHAKGIV+EK+G+EAKQP
Sbjct 76 KPRGLRTARKHVNHRRDQRWADKDYKKAHLGTRWKANPFGGASHAKGIVLEKVGVEAKQP 135
Query 67 NSAIRKCVRVQLIKNGKKITAFVPRDGCLNYIDENDEVLVAGFGRSGHAVGDIPGVRFKI 126
NSAIRKCVRVQLIKNGKKITAFVPRDG LNYI+ENDEVLVAGFGR GHAVGDIPGVRFK+
Sbjct 136 NSAIRKCVRVQLIKNGKKITAFVPRDGSLNYIEENDEVLVAGFGRKGHAVGDIPGVRFKV 195
Query 127 VKVSGVSLLALFKEKKEKPRS 147
VKV+ VSLLAL+KEKKE+PRS
Sbjct 196 VKVANVSLLALYKEKKERPRS 216
> Hs4506701
Length=143
Score = 247 bits (630), Expect = 7e-66, Method: Compositional matrix adjust.
Identities = 121/145 (83%), Positives = 133/145 (91%), Gaps = 2/145 (1%)
Query 3 MGACKPRGMRTARKLRNRRRVQKWADKQYKKAHLGTRWKANPFGGSSHAKGIVVEKLGIE 62
MG C RG+RTARKLR+ RR QKW DKQYKKAHLGT KANPFGG+SHAKGIV+EK+G+E
Sbjct 1 MGKC--RGLRTARKLRSHRRDQKWHDKQYKKAHLGTALKANPFGGASHAKGIVLEKVGVE 58
Query 63 AKQPNSAIRKCVRVQLIKNGKKITAFVPRDGCLNYIDENDEVLVAGFGRSGHAVGDIPGV 122
AKQPNSAIRKCVRVQLIKNGKKITAFVP DGCLN+I+ENDEVLVAGFGR GHAVGDIPGV
Sbjct 59 AKQPNSAIRKCVRVQLIKNGKKITAFVPNDGCLNFIEENDEVLVAGFGRKGHAVGDIPGV 118
Query 123 RFKIVKVSGVSLLALFKEKKEKPRS 147
RFK+VKV+ VSLLAL+K KKE+PRS
Sbjct 119 RFKVVKVANVSLLALYKGKKERPRS 143
> CE05747
Length=143
Score = 242 bits (618), Expect = 2e-64, Method: Compositional matrix adjust.
Identities = 113/141 (80%), Positives = 132/141 (93%), Gaps = 0/141 (0%)
Query 7 KPRGMRTARKLRNRRRVQKWADKQYKKAHLGTRWKANPFGGSSHAKGIVVEKLGIEAKQP 66
KP+G+ TARKL+ R+ Q+W DK+YKKAH+GTRWK+NPFGG+SHAKGIV+EK+G+EAKQP
Sbjct 3 KPKGLCTARKLKTHRQEQRWNDKRYKKAHIGTRWKSNPFGGASHAKGIVLEKIGVEAKQP 62
Query 67 NSAIRKCVRVQLIKNGKKITAFVPRDGCLNYIDENDEVLVAGFGRSGHAVGDIPGVRFKI 126
NSAIRKCVRVQLIKNGKKITAFVP DGCLN+++ENDEVLV+GFGRSGHAVGDIPGVRFKI
Sbjct 63 NSAIRKCVRVQLIKNGKKITAFVPNDGCLNFVEENDEVLVSGFGRSGHAVGDIPGVRFKI 122
Query 127 VKVSGVSLLALFKEKKEKPRS 147
VKV+ SL+ALFK KKE+PRS
Sbjct 123 VKVANTSLIALFKGKKERPRS 143
> At5g02960
Length=142
Score = 234 bits (598), Expect = 4e-62, Method: Compositional matrix adjust.
Identities = 116/141 (82%), Positives = 124/141 (87%), Gaps = 1/141 (0%)
Query 7 KPRGMRTARKLRNRRRVQKWADKQYKKAHLGTRWKANPFGGSSHAKGIVVEKLGIEAKQP 66
K RGM RKL+ R Q+WADKQYKK+HLG WK PF GSSHAKGIV+EK+GIEAKQP
Sbjct 3 KTRGMGAGRKLKRLRINQRWADKQYKKSHLGNEWK-KPFAGSSHAKGIVLEKIGIEAKQP 61
Query 67 NSAIRKCVRVQLIKNGKKITAFVPRDGCLNYIDENDEVLVAGFGRSGHAVGDIPGVRFKI 126
NSAIRKC RVQLIKNGKKI AFVP DGCLNYI+ENDEVL+AGFGR GHAVGDIPGVRFK+
Sbjct 62 NSAIRKCARVQLIKNGKKIAAFVPNDGCLNYIEENDEVLIAGFGRKGHAVGDIPGVRFKV 121
Query 127 VKVSGVSLLALFKEKKEKPRS 147
VKVSGVSLLALFKEKKEKPRS
Sbjct 122 VKVSGVSLLALFKEKKEKPRS 142
> YPR132w
Length=145
Score = 233 bits (594), Expect = 9e-62, Method: Compositional matrix adjust.
Identities = 114/145 (78%), Positives = 129/145 (88%), Gaps = 0/145 (0%)
Query 3 MGACKPRGMRTARKLRNRRRVQKWADKQYKKAHLGTRWKANPFGGSSHAKGIVVEKLGIE 62
MG KPRG+ +ARKLR RR +WA+ YKK LGT +K++PFGGSSHAKGIV+EKLGIE
Sbjct 1 MGKGKPRGLNSARKLRVHRRNNRWAENNYKKRLLGTAFKSSPFGGSSHAKGIVLEKLGIE 60
Query 63 AKQPNSAIRKCVRVQLIKNGKKITAFVPRDGCLNYIDENDEVLVAGFGRSGHAVGDIPGV 122
+KQPNSAIRKCVRVQLIKNGKK+TAFVP DGCLN++DENDEVL+AGFGR G A GDIPGV
Sbjct 61 SKQPNSAIRKCVRVQLIKNGKKVTAFVPNDGCLNFVDENDEVLLAGFGRKGKAKGDIPGV 120
Query 123 RFKIVKVSGVSLLALFKEKKEKPRS 147
RFK+VKVSGVSLLAL+KEKKEKPRS
Sbjct 121 RFKVVKVSGVSLLALWKEKKEKPRS 145
> YGR118w
Length=145
Score = 233 bits (594), Expect = 9e-62, Method: Compositional matrix adjust.
Identities = 114/145 (78%), Positives = 129/145 (88%), Gaps = 0/145 (0%)
Query 3 MGACKPRGMRTARKLRNRRRVQKWADKQYKKAHLGTRWKANPFGGSSHAKGIVVEKLGIE 62
MG KPRG+ +ARKLR RR +WA+ YKK LGT +K++PFGGSSHAKGIV+EKLGIE
Sbjct 1 MGKGKPRGLNSARKLRVHRRNNRWAENNYKKRLLGTAFKSSPFGGSSHAKGIVLEKLGIE 60
Query 63 AKQPNSAIRKCVRVQLIKNGKKITAFVPRDGCLNYIDENDEVLVAGFGRSGHAVGDIPGV 122
+KQPNSAIRKCVRVQLIKNGKK+TAFVP DGCLN++DENDEVL+AGFGR G A GDIPGV
Sbjct 61 SKQPNSAIRKCVRVQLIKNGKKVTAFVPNDGCLNFVDENDEVLLAGFGRKGKAKGDIPGV 120
Query 123 RFKIVKVSGVSLLALFKEKKEKPRS 147
RFK+VKVSGVSLLAL+KEKKEKPRS
Sbjct 121 RFKVVKVSGVSLLALWKEKKEKPRS 145
> SPBP4H10.13
Length=143
Score = 227 bits (579), Expect = 6e-60, Method: Compositional matrix adjust.
Identities = 108/141 (76%), Positives = 124/141 (87%), Gaps = 0/141 (0%)
Query 7 KPRGMRTARKLRNRRRVQKWADKQYKKAHLGTRWKANPFGGSSHAKGIVVEKLGIEAKQP 66
KP G+ ARKLRN RR ++WAD YKK LGT +K++PFGGSSHAKGIVVEK+G+EAKQP
Sbjct 3 KPAGLNAARKLRNHRREERWADAHYKKRLLGTAYKSSPFGGSSHAKGIVVEKIGVEAKQP 62
Query 67 NSAIRKCVRVQLIKNGKKITAFVPRDGCLNYIDENDEVLVAGFGRSGHAVGDIPGVRFKI 126
NSAIRKCVRVQLIKNGKK+TAFVP DGCLN++DENDEVL++GFGR G A GDIPGVRFK+
Sbjct 63 NSAIRKCVRVQLIKNGKKVTAFVPHDGCLNFVDENDEVLLSGFGRKGKAKGDIPGVRFKV 122
Query 127 VKVSGVSLLALFKEKKEKPRS 147
VKV+GV L ALF EKKEKPR+
Sbjct 123 VKVAGVGLSALFHEKKEKPRA 143
> SPAC23C11.02c
Length=143
Score = 227 bits (579), Expect = 6e-60, Method: Compositional matrix adjust.
Identities = 108/141 (76%), Positives = 124/141 (87%), Gaps = 0/141 (0%)
Query 7 KPRGMRTARKLRNRRRVQKWADKQYKKAHLGTRWKANPFGGSSHAKGIVVEKLGIEAKQP 66
KP G+ ARKLRN RR ++WAD YKK LGT +K++PFGGSSHAKGIVVEK+G+EAKQP
Sbjct 3 KPAGLNAARKLRNHRREERWADAHYKKRLLGTAYKSSPFGGSSHAKGIVVEKIGVEAKQP 62
Query 67 NSAIRKCVRVQLIKNGKKITAFVPRDGCLNYIDENDEVLVAGFGRSGHAVGDIPGVRFKI 126
NSAIRKCVRVQLIKNGKK+TAFVP DGCLN++DENDEVL++GFGR G A GDIPGVRFK+
Sbjct 63 NSAIRKCVRVQLIKNGKKVTAFVPHDGCLNFVDENDEVLLSGFGRKGKAKGDIPGVRFKV 122
Query 127 VKVSGVSLLALFKEKKEKPRS 147
VKV+GV L ALF EKKEKPR+
Sbjct 123 VKVAGVGLSALFHEKKEKPRA 143
> At3g09680
Length=136
Score = 220 bits (561), Expect = 7e-58, Method: Compositional matrix adjust.
Identities = 109/137 (79%), Positives = 118/137 (86%), Gaps = 1/137 (0%)
Query 11 MRTARKLRNRRRVQKWADKQYKKAHLGTRWKANPFGGSSHAKGIVVEKLGIEAKQPNSAI 70
M RKL+ R Q+WADK YKK++ G WK PF SSHAKGIV+EK+GIEAKQPNSAI
Sbjct 1 MGAGRKLKQLRITQRWADKHYKKSNRGNEWK-KPFACSSHAKGIVLEKIGIEAKQPNSAI 59
Query 71 RKCVRVQLIKNGKKITAFVPRDGCLNYIDENDEVLVAGFGRSGHAVGDIPGVRFKIVKVS 130
RKC RVQLIKNGKKI AFVP DGCLNYI+ENDEVL+AGFGR GHAVGDIPGVRFK+VKVS
Sbjct 60 RKCARVQLIKNGKKIAAFVPNDGCLNYIEENDEVLIAGFGRKGHAVGDIPGVRFKVVKVS 119
Query 131 GVSLLALFKEKKEKPRS 147
GVSLLALFKEKKEKPRS
Sbjct 120 GVSLLALFKEKKEKPRS 136
> ECU10g0400
Length=140
Score = 161 bits (407), Expect = 4e-40, Method: Compositional matrix adjust.
Identities = 79/136 (58%), Positives = 99/136 (72%), Gaps = 0/136 (0%)
Query 10 GMRTARKLRNRRRVQKWADKQYKKAHLGTRWKANPFGGSSHAKGIVVEKLGIEAKQPNSA 69
G+ +A L R+ ++ AD Y+K LGT++K + G + AK IV+EK+G+EAKQPNSA
Sbjct 3 GLFSANNLIKSRKAKRLADTAYRKRALGTKYKHSVLGRAPQAKAIVLEKIGVEAKQPNSA 62
Query 70 IRKCVRVQLIKNGKKITAFVPRDGCLNYIDENDEVLVAGFGRSGHAVGDIPGVRFKIVKV 129
IRK VR QLI GKKITAFVP DG + YI+ NDEV V GFG+ G +VGDIPG+RFK+ KV
Sbjct 63 IRKAVRCQLIATGKKITAFVPYDGSVTYIESNDEVTVEGFGKKGRSVGDIPGIRFKVCKV 122
Query 130 SGVSLLALFKEKKEKP 145
VSL A+F KKEKP
Sbjct 123 QNVSLHAIFTGKKEKP 138
> Hs22042004
Length=216
Score = 160 bits (405), Expect = 8e-40, Method: Compositional matrix adjust.
Identities = 83/116 (71%), Positives = 95/116 (81%), Gaps = 5/116 (4%)
Query 32 KKAHLGTRWKANPFGGSSHAKGIVVEKLGIEAKQPNSAIRKCVRVQLIKNGKKITAFVPR 91
KKAHLGT KANPFGG+S AKG+++EK+G+EAKQPNS RK VRVQ IKNGKKI AFVP
Sbjct 106 KKAHLGTAPKANPFGGASPAKGMMLEKVGVEAKQPNSTTRKSVRVQCIKNGKKIRAFVPN 165
Query 92 DGCLNYIDENDEVLVAGFGRSGHAVGDIPGVRFKIVKVSGVSLLALFKEKKEKPRS 147
DG +ENDE VAGFG+ HAVGDIPGV FK+VKV+ VSLLAL+K KKE+PRS
Sbjct 166 DG-----EENDEAPVAGFGQKSHAVGDIPGVCFKVVKVANVSLLALYKGKKERPRS 216
> Hs20470197
Length=130
Score = 74.7 bits (182), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 36/58 (62%), Positives = 46/58 (79%), Gaps = 0/58 (0%)
Query 34 AHLGTRWKANPFGGSSHAKGIVVEKLGIEAKQPNSAIRKCVRVQLIKNGKKITAFVPR 91
+ G+ KAN F G+S+AKGI++EK+ +EAKQPNSA RKC RVQLIKN KKIT+ V +
Sbjct 73 SDAGSALKANSFKGASNAKGIMLEKVEVEAKQPNSAFRKCFRVQLIKNSKKITSVVTQ 130
> 7290321
Length=140
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 42/109 (38%), Positives = 63/109 (57%), Gaps = 10/109 (9%)
Query 35 HLGTRWKANPFGGSSHAKGIVVEKLGIEAKQPNSAIRKCVRVQLIKNGKKITAFVPRDGC 94
H+ TR P G AKG+V++ L + K+PNSA RKCV V+L GK++ A++P G
Sbjct 42 HIKTRPPRQPLDGKPFAKGVVLKTLIKKPKKPNSANRKCVLVRL-STGKEMVAYIPGIG- 99
Query 95 LNYIDENDEVLVAGFGRSGHAVGDIPGVRFKIVKVSGVSLLALFKEKKE 143
+ + E++ VL GR + D+PGV+ K V+ GV LA +K +
Sbjct 100 -HNLQEHNIVL-CRVGR----LQDVPGVKLKAVR--GVYDLAHVVKKSQ 140
> Hs11056056
Length=138
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 33/88 (37%), Positives = 50/88 (56%), Gaps = 8/88 (9%)
Query 41 KANPFGGSSHAKGIVVEKLGIEAKQPNSAIRKCVRVQLIKNGKKITAFVPRDGCLNYIDE 100
K P G KG+V+ + K+PNSA RKC RV+L G++ F+P +G + + E
Sbjct 48 KLGPTEGRPQLKGVVLCTFTRKPKKPNSANRKCCRVRL-STGREAVCFIPGEG--HTLQE 104
Query 101 NDEVLVAGFGRSGHAVGDIPGVRFKIVK 128
+ VLV G GR+ D+PGV+ +V+
Sbjct 105 HQIVLVEG-GRT----QDLPGVKLTVVR 127
> AtCh002
Length=123
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 35/88 (39%), Positives = 50/88 (56%), Gaps = 8/88 (9%)
Query 41 KANPFGGSSHAKGIVVEKLGIEAKQPNSAIRKCVRVQLIKNGKKITAFVPRDGCLNYIDE 100
K+ G +G I K+PNSA+RK RV+L +G +ITA++P G + + E
Sbjct 20 KSPALRGCPQRRGTCTRVYTITPKKPNSALRKVARVRLT-SGFEITAYIP--GIGHNLQE 76
Query 101 NDEVLVAGFGRSGHAVGDIPGVRFKIVK 128
+ VLV G GR V D+PGVR+ IV+
Sbjct 77 HSVVLVRG-GR----VKDLPGVRYHIVR 99
> AtCh001
Length=123
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 35/88 (39%), Positives = 50/88 (56%), Gaps = 8/88 (9%)
Query 41 KANPFGGSSHAKGIVVEKLGIEAKQPNSAIRKCVRVQLIKNGKKITAFVPRDGCLNYIDE 100
K+ G +G I K+PNSA+RK RV+L +G +ITA++P G + + E
Sbjct 20 KSPALRGCPQRRGTCTRVYTITPKKPNSALRKVARVRLT-SGFEITAYIP--GIGHNLQE 76
Query 101 NDEVLVAGFGRSGHAVGDIPGVRFKIVK 128
+ VLV G GR V D+PGVR+ IV+
Sbjct 77 HSVVLVRG-GR----VKDLPGVRYHIVR 99
> YNR036c
Length=153
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 50/80 (62%), Gaps = 8/80 (10%)
Query 52 KGIVVEKLGIEAKQPNSAIRKCVRVQLIKNGKKITAFVPRDGCLNYIDENDEVLVAGFGR 111
KG+V+ + ++ K+PNSA RK RV+L NG ++A++P +G + E+ V V G GR
Sbjct 59 KGVVLRVMVLKPKKPNSAQRKACRVRLT-NGNVVSAYIPGEG--HDAQEHSIVYVRG-GR 114
Query 112 SGHAVGDIPGVRFKIVKVSG 131
D+PGV++ +++ +G
Sbjct 115 ----CQDLPGVKYHVIRGAG 130
> SPAC4F8.06
Length=146
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 29/82 (35%), Positives = 48/82 (58%), Gaps = 8/82 (9%)
Query 47 GSSHAKGIVVEKLGIEAKQPNSAIRKCVRVQLIKNGKKITAFVPRDGCLNYIDENDEVLV 106
GS +G+ ++ K+PNSA+RK RV+L G+ +TA++P G + E+ VL+
Sbjct 47 GSPFRRGVCTRVFTVKPKKPNSAVRKVARVRL-STGRSVTAYIP--GIGHNAQEHAVVLL 103
Query 107 AGFGRSGHAVGDIPGVRFKIVK 128
G GR+ D PGV++ +V+
Sbjct 104 RG-GRA----QDCPGVQYHVVR 120
> CE18920
Length=157
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 38/62 (61%), Gaps = 3/62 (4%)
Query 47 GSSHAKGIVVEKLGIEAKQPNSAIRKCVRVQLIKNGKKITAFVPRDGCLNYIDENDEVLV 106
G SH KGIV++ + K+PNS RKC V+L G ++ A++P G + + E+ +VLV
Sbjct 72 GYSHYKGIVLKTVIRHPKKPNSGNRKCAIVRL-STGAEVCAYIPNVG--HNLQEHSQVLV 128
Query 107 AG 108
G
Sbjct 129 KG 130
> AtMi080
Length=125
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 41/65 (63%), Gaps = 8/65 (12%)
Query 64 KQPNSAIRKCVRVQLIKNGKKITAFVPRDGCLNYIDENDEVLVAGFGRSGHAVGDIPGVR 123
K+PNSA+RK +V+L N I A++P +G + + E+ VL+ G GR V D+PGV+
Sbjct 43 KKPNSALRKIAKVRL-SNRHDIFAYIPGEG--HNLQEHSTVLIRG-GR----VKDLPGVK 94
Query 124 FKIVK 128
F ++
Sbjct 95 FHCIR 99
> Hs20559030
Length=311
Score = 31.2 bits (69), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 14/63 (22%), Positives = 32/63 (50%), Gaps = 0/63 (0%)
Query 77 QLIKNGKKITAFVPRDGCLNYIDENDEVLVAGFGRSGHAVGDIPGVRFKIVKVSGVSLLA 136
Q+++ + ++ F+P G + +E +E + F +G+ + D+PG ++L
Sbjct 14 QVVETAQHLSPFIPTSGAFSGKNEAEERVALMFPGAGNHLDDLPGPLGSRSSTDHSNVLD 73
Query 137 LFK 139
L+K
Sbjct 74 LWK 76
> CE18959
Length=811
Score = 30.8 bits (68), Expect = 0.99, Method: Composition-based stats.
Identities = 12/16 (75%), Positives = 13/16 (81%), Gaps = 0/16 (0%)
Query 86 TAFVPRDGCLNYIDEN 101
T FVP+DG LN IDEN
Sbjct 653 TPFVPKDGVLNVIDEN 668
> At2g07675
Length=125
Score = 30.4 bits (67), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 36/65 (55%), Gaps = 8/65 (12%)
Query 64 KQPNSAIRKCVRVQLIKNGKKITAFVPRDGCLNYIDENDEVLVAGFGRSGHAVGDIPGVR 123
K+PNSA RK +V+L N I A +P +G + E+ VL+ G GR V D PGV+
Sbjct 43 KKPNSAPRKIAKVRL-SNRHDIFAHIPGEG--HNSQEHSTVLIRG-GR----VKDSPGVK 94
Query 124 FKIVK 128
++
Sbjct 95 SHCIR 99
> At5g10100
Length=372
Score = 28.9 bits (63), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 22/89 (24%), Positives = 40/89 (44%), Gaps = 9/89 (10%)
Query 21 RRVQKWADKQYKKAHLGTRWKANPFGGSSHAKGIVVEKLGIEAKQPNSAIRKCVRVQLIK 80
+R+ W D +A T K+ P S+ ++L Q SA+ K ++
Sbjct 57 QRINAWVDSM--RASSPTHLKSLPSSISTQ------QQLNSWIMQHPSALEKFEQIMEAS 108
Query 81 NGKKITAFVPRDGCLN-YIDENDEVLVAG 108
GK+I F+ DG L+ +D+ D+ ++
Sbjct 109 RGKQIVMFLDYDGTLSPIVDDPDKAFMSS 137
> Hs18544711
Length=468
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 40 WKANPFGGSSHAKGIVVEKLGIEAKQPNSAIRKCVRVQLIKNG 82
W+ S A GI + L +A++P+SA ++C R++L +G
Sbjct 351 WEIEVGDKSEWAIGICKDSLPTKARRPSSAQQECWRIELQDDG 393
Lambda K H
0.320 0.136 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1785281974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40