bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2704_orf1
Length=182
Score E
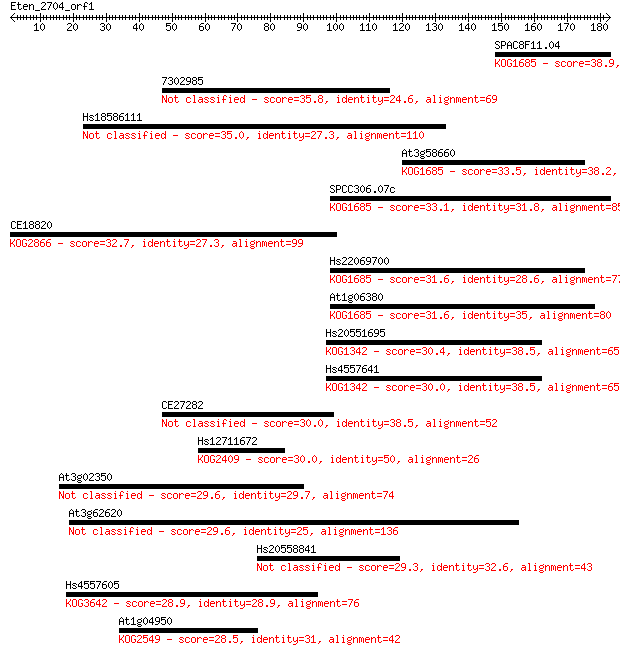
Sequences producing significant alignments: (Bits) Value
SPAC8F11.04 38.9 0.006
7302985 35.8 0.047
Hs18586111 35.0 0.080
At3g58660 33.5 0.23
SPCC306.07c 33.1 0.35
CE18820 32.7 0.44
Hs22069700 31.6 0.96
At1g06380 31.6 0.99
Hs20551695 30.4 2.2
Hs4557641 30.0 2.5
CE27282 30.0 2.7
Hs12711672 30.0 3.0
At3g02350 29.6 3.0
At3g62620 29.6 3.2
Hs20558841 29.3 4.9
Hs4557605 28.9 6.6
At1g04950 28.5 8.3
> SPAC8F11.04
Length=373
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 26/35 (74%), Gaps = 0/35 (0%)
Query 148 QRIYPHLIELPHSPLASNSEVCLIVRDPQRKWKDL 182
+++ P+ I + + + S+SE CLIV+DPQR +KDL
Sbjct 67 RKLIPYKIAIKNPVIPSSSEACLIVKDPQRVYKDL 101
> 7302985
Length=203
Score = 35.8 bits (81), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 17/69 (24%), Positives = 36/69 (52%), Gaps = 2/69 (2%)
Query 47 RSSSAPQSLSSPSQPRKKAKSTPTTAEKAKGCPQAPAASAPTVIKIGSGGVVDCQALEKA 106
++ + + SPS+ K S P E + C + AS +++KIG + DC++++
Sbjct 55 KNGGGVRFVESPSRLWKDYNSLP--GENTQFCIKVVDASNSSIMKIGLEHLKDCRSIDTV 112
Query 107 VKALCRHVQ 115
+ C+H++
Sbjct 113 IFHNCKHLE 121
> Hs18586111
Length=480
Score = 35.0 bits (79), Expect = 0.080, Method: Compositional matrix adjust.
Identities = 30/116 (25%), Positives = 49/116 (42%), Gaps = 9/116 (7%)
Query 23 RKNGIAAAAKPLSVDTPDGKHKAKRSSSAPQSLSSPSQPRKKAK-STPTTAEKAKGCPQA 81
+ G+ K V+ G K +R + Q L S K+ + STP G
Sbjct 186 QDQGLTVQEKDRDVEASWGGFKNRRMDVSCQRLDRLSVHLKEIQGSTPFRHWCEMG---Q 242
Query 82 PAASAPTVIKIGSGGVVDCQALEKAVKA-----LCRHVQKLQEKEEVPDLLAMNSG 132
A P V+ G+GG+ C+ AV LCRH++ Q E++ ++++G
Sbjct 243 KGAKEPAVVSAGAGGLAGCKVDSAAVLWIWCPRLCRHIEGTQGSEQMARCSSVHAG 298
> At3g58660
Length=446
Score = 33.5 bits (75), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 29/58 (50%), Gaps = 5/58 (8%)
Query 120 KEEVPDLLAMNSGPTVSLMFSTQNIPVKQRIYPHLIELPH---SPLASNSEVCLIVRD 174
K E P LL + V L + + IP K R + I LPH +P + E+CLI+ D
Sbjct 45 KTEKPQLLEEDE--LVYLFVTLKKIPQKTRTNAYRIPLPHPLINPTVDSPEICLIIDD 100
> SPCC306.07c
Length=284
Score = 33.1 bits (74), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 27/90 (30%), Positives = 48/90 (53%), Gaps = 19/90 (21%)
Query 98 VDCQALEKAVKALCRHVQ---KLQEKEEVPDLLAMNSGPTVSLMFSTQNIPVKQRIY--P 152
+D + LEK ++AL +H++ K EKE+V + +N+ PV++ P
Sbjct 17 IDIKLLEKTIRALLQHIRSSDKPIEKEKV--YIQVNTFQ-----------PVEKESLRRP 63
Query 153 HLIELPHSPLASNSEVCLIVRDPQRKWKDL 182
+ LPH + ++ CLIV+D Q+ ++DL
Sbjct 64 SKVFLPHR-IMHVTDACLIVKDSQQTYQDL 92
> CE18820
Length=459
Score = 32.7 bits (73), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 45/99 (45%), Gaps = 11/99 (11%)
Query 1 TGRCRLFGCFISNEVTMIKVKKRKNGIAAAAKPLSVDTPDGKHKAKRSSSAPQSLSSPSQ 60
T +CR N ++KK+ + +A K +VD+ K K+ SA SL S+
Sbjct 102 TVQCR------ENHAVAEELKKQISELAHIRKSETVDS-----KIKKLESAVTSLREQSK 150
Query 61 PRKKAKSTPTTAEKAKGCPQAPAASAPTVIKIGSGGVVD 99
P KKA +T + + + T+++I G V+D
Sbjct 151 PLKKALNTALYSTDTELLELVGQTTRETLLEINGGAVID 189
> Hs22069700
Length=296
Score = 31.6 bits (70), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 38/78 (48%), Gaps = 8/78 (10%)
Query 98 VDCQALEKAVKALCRHVQKLQEKEEVPDLLAMNSGPTVSLMFSTQNIPVKQ-RIYPHLIE 156
+D + + KAV AL H + + L +N ++ LM IP K+ R+ +
Sbjct 31 LDKEQVRKAVDALLTHCKSRKNNYG----LLLNENESLFLMVVLWKIPSKELRVR---LT 83
Query 157 LPHSPLASNSEVCLIVRD 174
LPHS + + ++CL +D
Sbjct 84 LPHSIRSDSEDICLFTKD 101
> At1g06380
Length=254
Score = 31.6 bits (70), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 40/85 (47%), Gaps = 9/85 (10%)
Query 98 VDCQALEKAVKALCRHVQKLQEKEEVPDLLAMNSGPTVSLMFSTQNIPVKQRIYPHLIEL 157
VD Q + +AVK+L + + E L N G V L+ + + IP R P +I L
Sbjct 18 VDPQNVNRAVKSLLKWWDSKSKTENSESL--ENDG-FVYLIVTLKRIPQLDRTNPLMIPL 74
Query 158 PHSPLASNS-----EVCLIVRDPQR 177
PH PL E+CLI+ D +
Sbjct 75 PH-PLIDLVAEDPPELCLIIDDKHK 98
> Hs20551695
Length=488
Score = 30.4 bits (67), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 32/80 (40%), Gaps = 25/80 (31%)
Query 97 VVDCQALEKAVKALCRHVQKLQEKEEVPDLLAMNS-----GPTVSLMFS-----TQNIP- 145
V C E AV C E+P+ L N GP L S QN P
Sbjct 309 VARCWTYETAVALDC----------EIPNELPYNDYFEYFGPDFKLHISPSNMTNQNTPE 358
Query 146 ----VKQRIYPHLIELPHSP 161
+KQR++ +L LPH+P
Sbjct 359 YMEKIKQRLFENLRMLPHAP 378
> Hs4557641
Length=488
Score = 30.0 bits (66), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 32/80 (40%), Gaps = 25/80 (31%)
Query 97 VVDCQALEKAVKALCRHVQKLQEKEEVPDLLAMNS-----GPTVSLMFS-----TQNIP- 145
V C E AV C E+P+ L N GP L S QN P
Sbjct 309 VARCWTYETAVALDC----------EIPNELPYNDYFEYFGPDFKLHISPSNMTNQNTPE 358
Query 146 ----VKQRIYPHLIELPHSP 161
+KQR++ +L LPH+P
Sbjct 359 YMEKIKQRLFENLRMLPHAP 378
> CE27282
Length=789
Score = 30.0 bits (66), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 27/52 (51%), Gaps = 2/52 (3%)
Query 47 RSSSAPQSLSSPSQPRKKAKSTPTTAEKAKGCPQAPAASAPTVIKIGSGGVV 98
R +S +S PS RKK STPT + PQ P+ SA TV + +G +
Sbjct 16 RGNSGARSHKEPSAKRKKPTSTPTV--RCGTVPQEPSTSAATVTQKPAGSTM 65
> Hs12711672
Length=591
Score = 30.0 bits (66), Expect = 3.0, Method: Composition-based stats.
Identities = 13/26 (50%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 58 PSQPRKKAKSTPTTAEKAKGCPQAPA 83
PSQPRKK + P T +K + P A A
Sbjct 348 PSQPRKKKREAPLTGKKKRKSPFAAA 373
> At3g02350
Length=561
Score = 29.6 bits (65), Expect = 3.0, Method: Composition-based stats.
Identities = 22/81 (27%), Positives = 37/81 (45%), Gaps = 15/81 (18%)
Query 16 TMIKVKKRKNGIAAAAKPLSVDTPDGKHKAKRSSSAPQSLSSPSQPR-------KKAKST 68
T +K++K K+ I A + L+ KAK++ + +S+ S P+ +
Sbjct 177 TQLKIQKLKDTIFAVQEQLT--------KAKKNGAVASLISAKSVPKSLHCLAMRLVGER 228
Query 69 PTTAEKAKGCPQAPAASAPTV 89
+ EK K P PAA PT+
Sbjct 229 ISNPEKYKDAPPDPAAEDPTL 249
> At3g62620
Length=429
Score = 29.6 bits (65), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 34/139 (24%), Positives = 59/139 (42%), Gaps = 14/139 (10%)
Query 19 KVKKRKNGIAAAAKPLSVDTPDGK---HKAKRSSSAPQSLSSPSQPRKKAKSTPTTAEKA 75
KV +K A A+ P + D D K K +SSS Q S SQ + S P +
Sbjct 231 KVNGKKKTEANASSP-AKDATDSKVSFSKPTKSSSQVQDQKSKSQKKISPSSEPENETIS 289
Query 76 KGCPQAPAASAPTVIKIGSGGVVDCQALEKAVKALCRHVQKLQEKEEVPDLLAMNSGPTV 135
KG + S+ VI+IG+ ++ + + + ++C + L+ V + TV
Sbjct 290 KGSATSNTLSSVLVIRIGN---LNSKTADSLIHSMCFSIGPLEGLTRVNE-------DTV 339
Query 136 SLMFSTQNIPVKQRIYPHL 154
++F +++ I L
Sbjct 340 DVLFRLKDLKEADSILEEL 358
> Hs20558841
Length=118
Score = 29.3 bits (64), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 22/43 (51%), Gaps = 1/43 (2%)
Query 76 KGCPQAPAASAPTVIKIGSGGVVDCQALEKAVKALCRHVQKLQ 118
+G P + P+V+ + S G+ C EK + A C HV +L
Sbjct 39 QGSPMKDRLNLPSVLVLNSCGIT-CAGDEKEIAAFCAHVSELD 80
> Hs4557605
Length=554
Score = 28.9 bits (63), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 22/81 (27%), Positives = 34/81 (41%), Gaps = 7/81 (8%)
Query 18 IKVKKRKNGIAAAAKPLSVDTPDGKHKAKRSSSAPQSLSSPSQPRKKAKSTPTTAEKAKG 77
+ ++KR N + + + T G H +K S+ + SS PR S+P +A
Sbjct 384 LNMRKRTNALVHSESDVGNRTEVGNHSSKSSTVVQE--SSKGTPRSYLASSPNPFSRANA 441
Query 78 CPQAPAASA-----PTVIKIG 93
AA A PT I+ G
Sbjct 442 AETISAARALPSASPTSIRTG 462
> At1g04950
Length=549
Score = 28.5 bits (62), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 13/44 (29%), Positives = 27/44 (61%), Gaps = 2/44 (4%)
Query 34 LSVDTPDGKHKAKRSSSAPQSLSSPSQPRKKAKST--PTTAEKA 75
+++D PDG H +S SAP + +P + +++ P+++E+A
Sbjct 440 ITMDGPDGVHSQDQSGSAPMQVDNPVENDNPPQNSVQPSSSEQA 483
Lambda K H
0.314 0.128 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2878611680
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40