bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
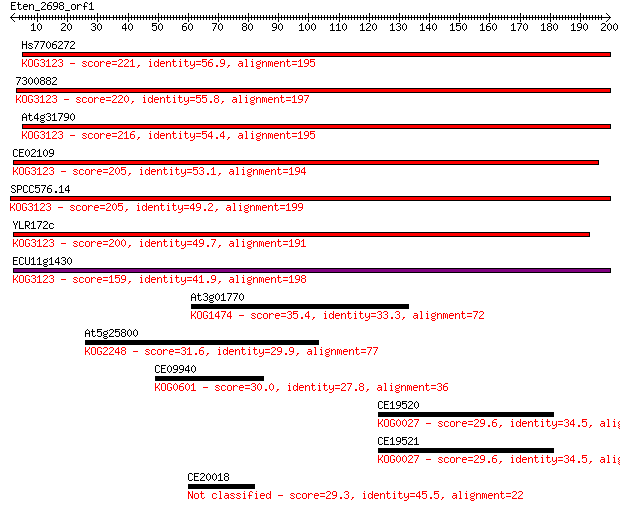
Query= Eten_2698_orf1
Length=199
Score E
Sequences producing significant alignments: (Bits) Value
Hs7706272 221 6e-58
7300882 220 1e-57
At4g31790 216 2e-56
CE02109 205 4e-53
SPCC576.14 205 4e-53
YLR172c 200 1e-51
ECU11g1430 159 3e-39
At3g01770 35.4 0.068
At5g25800 31.6 0.99
CE09940 30.0 2.8
CE19520 29.6 3.8
CE19521 29.6 3.9
CE20018 29.3 5.0
> Hs7706272
Length=297
Score = 221 bits (564), Expect = 6e-58, Method: Compositional matrix adjust.
Identities = 111/212 (52%), Positives = 144/212 (67%), Gaps = 17/212 (8%)
Query 5 MLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQLYRF 64
+L+ A VA LVVGDPF ATTHSDL LRA G+ +VIHNASIM+AV CGLQLY+F
Sbjct 68 ILKDADISDVAFLVVGDPFGATTHSDLVLRATKLGIPYRVIHNASIMNAVGCCGLQLYKF 127
Query 65 GETVSVPFFD------------GGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENL 112
GETVS+ W+P SF++K++KNR+N MHTLCLLDIKVKEQ++ENL
Sbjct 128 GETVSIMLISVMLHSLWLVIHLDTWRPESFFDKVKKNRQNGMHTLCLLDIKVKEQSLENL 187
Query 113 MKGNNIFEPPRFMTVNTAIRQLFEAAEMQ---GDEGVAS--ILAFGLARVGADDQAIVSG 167
+KG I+EPPR+M+VN A +QL E + Q G+E + L GLARVGADDQ I +G
Sbjct 188 IKGRKIYEPPRYMSVNQAAQQLLEIVQNQRIRGEEPAVTEETLCVGLARVGADDQKIAAG 247
Query 168 PLQELLNADLGGPLHSLVLCAPELHEIEQKFV 199
L+++ DLG PLHSL++ +H +E + +
Sbjct 248 TLRQMCTVDLGEPLHSLIITGGSIHPMEMEML 279
> 7300882
Length=558
Score = 220 bits (561), Expect = 1e-57, Method: Compositional matrix adjust.
Identities = 110/202 (54%), Positives = 139/202 (68%), Gaps = 5/202 (2%)
Query 3 EAMLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQLY 62
+ +L A VA LVVGDPF ATTH+D LRA++K + KVIHNASIM+AV CGLQLY
Sbjct 343 DEILAGAGESDVALLVVGDPFGATTHTDFILRAKEKNIPYKVIHNASIMNAVGCCGLQLY 402
Query 63 RFGETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNIFEPP 122
+FGETVS+P++D WKP SFY+KI+ NR +NMHTLCLLDIKVKE T E+LM+ + PP
Sbjct 403 KFGETVSIPYWDETWKPDSFYDKIKLNRLHNMHTLCLLDIKVKEPTPESLMRKRKEYMPP 462
Query 123 RFMTVNTAIRQLFEAAEMQGDEGVASI-----LAFGLARVGADDQAIVSGPLQELLNADL 177
RFMTV A QL E + ++ L GLARVG + Q I G L E+ + DL
Sbjct 463 RFMTVAEAAHQLLSIVEKKDSLEKNTVLNEQSLCVGLARVGQESQQIAVGTLLEMRSTDL 522
Query 178 GGPLHSLVLCAPELHEIEQKFV 199
GGPLHSL++ A E+H +E +F+
Sbjct 523 GGPLHSLIIPAKEMHPLEVEFL 544
> At4g31790
Length=277
Score = 216 bits (551), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 106/197 (53%), Positives = 139/197 (70%), Gaps = 3/197 (1%)
Query 5 MLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQLYRF 64
M++ A VA LVVGDPF ATTHSDL +RA+ GV V+V+HNAS+M+AV CGLQLY +
Sbjct 73 MIDEAIDNDVAFLVVGDPFGATTHSDLVVRAKTLGVKVEVVHNASVMNAVGICGLQLYHY 132
Query 65 GETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNI-FEPPR 123
GETVS+PFF W+P SFYEKI+KNR +HTLCLLDI+VKE T E+L +G +EPPR
Sbjct 133 GETVSIPFFTETWRPDSFYEKIKKNRSLGLHTLCLLDIRVKEPTFESLCRGGKKQYEPPR 192
Query 124 FMTVNTAIRQLFEAAEMQGDEGVA-SILAFGLARVGADDQAIVSGPLQELLNADLGGPLH 182
+M+VNTAI QL E + GD G AR+G++DQ IV+G +++L + D G PLH
Sbjct 193 YMSVNTAIEQLLEVEQKHGDSVYGEDTQCVGFARLGSEDQTIVAGTMKQLESVDFGAPLH 252
Query 183 SLVLCAPELHEIEQKFV 199
LV+ E H +E++ +
Sbjct 253 CLVIVG-ETHPVEEEML 268
> CE02109
Length=274
Score = 205 bits (522), Expect = 4e-53, Method: Compositional matrix adjust.
Identities = 103/199 (51%), Positives = 138/199 (69%), Gaps = 6/199 (3%)
Query 2 TEAMLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQL 61
++A+L A + VA LVVGDPF ATTH+DL LRA+ + + VKVIHNASIM+AV CGLQL
Sbjct 68 SDAILNGADKEDVALLVVGDPFGATTHADLVLRAKQQNIPVKVIHNASIMNAVGCCGLQL 127
Query 62 YRFGETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNIFEP 121
Y FGETVS+ + W+P S+Y+KI NR+ MHTLCLLDIK KEQTVEN+M+G IFEP
Sbjct 128 YNFGETVSIVMWTDEWQPESYYDKIALNRKRGMHTLCLLDIKTKEQTVENMMRGRKIFEP 187
Query 122 PRFMTVNTAIRQL---FEAAEMQGDEGV--ASILAFGLARVGADDQAIVSGPLQELLNAD 176
R+ + A RQL +E + +G+E + + GLARVG D+Q IV ++++ +
Sbjct 188 ARYQKCSEAARQLLTIYERRKAKGEECAYDENTMVVGLARVGWDNQKIVYASMKDMSEME 247
Query 177 LGGPLHSLVLCAPELHEIE 195
+G PLHSL++ E H +E
Sbjct 248 MGEPLHSLII-PGETHPLE 265
> SPCC576.14
Length=283
Score = 205 bits (522), Expect = 4e-53, Method: Compositional matrix adjust.
Identities = 98/200 (49%), Positives = 142/200 (71%), Gaps = 1/200 (0%)
Query 1 STEAMLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQ 60
S++ +L+ A VA LVVGDP ATTH+DL +RAR+ + V++IHNASIM+A+ ACGLQ
Sbjct 64 SSDEILKDADNCDVAMLVVGDPMGATTHADLVIRARELKIPVRMIHNASIMNAIGACGLQ 123
Query 61 LYRFGETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNIFE 120
LY+FG+TVS+ FF+ ++P SFY+ I++N +HTL LLDIKVKEQ+ ENL +G ++E
Sbjct 124 LYKFGQTVSLVFFENNYRPQSFYDHIKENVSLGLHTLVLLDIKVKEQSWENLARGRKVYE 183
Query 121 PPRFMTVNTAIRQLFEAAE-MQGDEGVASILAFGLARVGADDQAIVSGPLQELLNADLGG 179
PPR+M+ + A +Q+ E E Q + L + R+G+DDQ I +G LQEL D+G
Sbjct 184 PPRYMSASLAAQQMLEVEEDRQENICTPDSLCVAVGRMGSDDQVIFAGTLQELAEHDIGP 243
Query 180 PLHSLVLCAPELHEIEQKFV 199
PLHS+VL ++H++E +F+
Sbjct 244 PLHSVVLVGRDVHDLELEFL 263
> YLR172c
Length=300
Score = 200 bits (509), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 95/192 (49%), Positives = 133/192 (69%), Gaps = 1/192 (0%)
Query 2 TEAMLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQL 61
++ +L A + VA LVVGDPF ATTH+DL LRA+ + + V++IHNAS+M+AV ACGLQL
Sbjct 66 SKQILNNADKEDVAFLVVGDPFGATTHTDLVLRAKREAIPVEIIHNASVMNAVGACGLQL 125
Query 62 YRFGETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNIFEP 121
Y FG+TVS+ FF W+P S+Y+KI +NR+ +HTL LLDIKVKEQ++EN+ +G I+EP
Sbjct 126 YNFGQTVSMVFFTDNWRPDSWYDKIWENRKIGLHTLVLLDIKVKEQSIENMARGRLIYEP 185
Query 122 PRFMTVNTAIRQLFEAAEMQGDEGVA-SILAFGLARVGADDQAIVSGPLQELLNADLGGP 180
PR+M++ QL E E +G + A ++R+G+ Q+ SG + EL N D G P
Sbjct 186 PRYMSIAQCCEQLLEIEEKRGTKAYTPDTPAVAISRLGSSSQSFKSGTISELANYDSGEP 245
Query 181 LHSLVLCAPELH 192
LHSLV+ + H
Sbjct 246 LHSLVILGRQCH 257
> ECU11g1430
Length=262
Score = 159 bits (402), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 83/199 (41%), Positives = 120/199 (60%), Gaps = 17/199 (8%)
Query 2 TEAMLEAAKTKRVACLVVGDPFCATTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQL 61
T+ +++ + + V+ LVVG P ATTHSD+ +RA++KGV V+VIHNASI++ + CGL
Sbjct 75 TDKIVDESCRENVSLLVVGTPLFATTHSDIMIRAKEKGVDVEVIHNASIINVLGCCGLYS 134
Query 62 YRFGETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVENLMKGNNIFEP 121
Y FG VS+P+F WKP SFY+ I +N ++N+HTLCLLDI+ E
Sbjct 135 YSFGRVVSIPYFTERWKPTSFYDNIVRNHQSNLHTLCLLDIRTDED-------------- 180
Query 122 PRFMTVNTAIRQLFEAAEMQGDEGV-ASILAFGLARVGADDQAIVSGPLQELLNADLGGP 180
RFM+VN A+ Q+ EAA + G + F + R G+ + IV G + +L G P
Sbjct 181 -RFMSVNEAVDQILEAAAITGSPLINEDTRIFAVCRFGSPSEEIVYGKIGDLKLRSFGDP 239
Query 181 LHSLVLCAPELHEIEQKFV 199
LHSL++ A EL +E + V
Sbjct 240 LHSLIVPA-ELDRVEAELV 257
> At3g01770
Length=601
Score = 35.4 bits (80), Expect = 0.068, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 39/77 (50%), Gaps = 6/77 (7%)
Query 61 LYRFGETVSVPFFDGGWKPVSFYEKIQKNRENNMHTLCLLDIKVKEQTVE----NLMKGN 116
+YRF +T+S FF+ WK + K+ +N+ TL DI + E + N +K N
Sbjct 212 VYRFADTLSK-FFEVRWKTIEKKSSGTKSEPSNLATLAHKDIAIPEPVAKKRKMNAVKRN 270
Query 117 NIFEP-PRFMTVNTAIR 132
++ EP R MT ++
Sbjct 271 SLLEPAKRVMTDEDRVK 287
> At5g25800
Length=534
Score = 31.6 bits (70), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 34/78 (43%), Gaps = 7/78 (8%)
Query 26 TTHSDLYLRARDKGVTVKVIHNASIMSAVAACGLQLYRFGE-TVSVPFFDGGWKPVSFYE 84
T + L + R K + ++S+ AC L F E T +PF PVS+Y
Sbjct 129 TIDTILTCKGRKKKTVTSSVEPPPLVSSPEACNLMGKSFVELTKDIPF------PVSYYT 182
Query 85 KIQKNRENNMHTLCLLDI 102
QK E N +T L++
Sbjct 183 LSQKEMEQNGYTFEKLEL 200
> CE09940
Length=468
Score = 30.0 bits (66), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 10/36 (27%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 49 SIMSAVAACGLQLYRFGETVSVPFFDGGWKPVSFYE 84
SI S + + G+ + G + +P + GW+P+ +E
Sbjct 281 SIFSDIFSFGISILEVGTNIHLPSYGTGWEPIRKWE 316
> CE19520
Length=567
Score = 29.6 bits (65), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 33/62 (53%), Gaps = 5/62 (8%)
Query 123 RFMTVNTAIRQLFEAAEMQGDEGV----ASILAFGLARVGADDQAIVSGPLQELLNADLG 178
R MT+ A RQ+ + A++ GD + A +AF +GA D A + G + E ++ +L
Sbjct 186 RGMTIRLA-RQIMKNADLNGDGHISVDEAQAIAFEQEGIGAGDVASMVGSVDENMDGELN 244
Query 179 GP 180
P
Sbjct 245 AP 246
> CE19521
Length=482
Score = 29.6 bits (65), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 33/62 (53%), Gaps = 5/62 (8%)
Query 123 RFMTVNTAIRQLFEAAEMQGDEGV----ASILAFGLARVGADDQAIVSGPLQELLNADLG 178
R MT+ A RQ+ + A++ GD + A +AF +GA D A + G + E ++ +L
Sbjct 186 RGMTIRLA-RQIMKNADLNGDGHISVDEAQAIAFEQEGIGAGDVASMVGSVDENMDGELN 244
Query 179 GP 180
P
Sbjct 245 AP 246
> CE20018
Length=329
Score = 29.3 bits (64), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 10/22 (45%), Positives = 16/22 (72%), Gaps = 0/22 (0%)
Query 60 QLYRFGETVSVPFFDGGWKPVS 81
+L ++G T S+PF++ WKP S
Sbjct 3 RLIQYGNTDSIPFYNCSWKPQS 24
Lambda K H
0.321 0.135 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3443493696
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40