bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
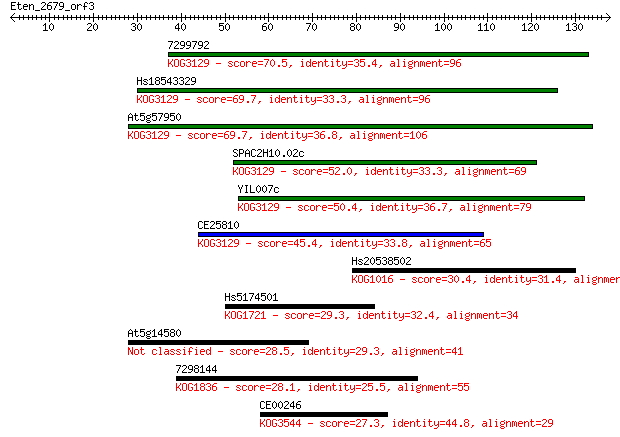
Query= Eten_2679_orf3
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
7299792 70.5 8e-13
Hs18543329 69.7 2e-12
At5g57950 69.7 2e-12
SPAC2H10.02c 52.0 3e-07
YIL007c 50.4 1e-06
CE25810 45.4 3e-05
Hs20538502 30.4 0.94
Hs5174501 29.3 2.6
At5g14580 28.5 4.2
7298144 28.1 4.5
CE00246 27.3 7.9
> 7299792
Length=220
Score = 70.5 bits (171), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 34/96 (35%), Positives = 53/96 (55%), Gaps = 0/96 (0%)
Query 37 AGASSLAAQAEAVYRERQQIEEKMEALASFLTAEGMPGLKGPLVDPEGFPRADIDVHAVL 96
A ++ + E + ++Q+E ++ L A G+ GPLVD EGFPR DIDV+ V
Sbjct 2 AAGTTTKERLERLINAKKQLEAQINRNGQILAANDNVGMSGPLVDAEGFPRNDIDVYQVR 61
Query 97 QARQQLACLKTDHREVQQRLEETLLLLHAANAKDKP 132
ARQ + CL+ DH+E+ +++ L H+ A P
Sbjct 62 LARQTIICLQNDHKELMNQIQTLLNQYHSEIATTDP 97
> Hs18543329
Length=223
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 57/96 (59%), Gaps = 0/96 (0%)
Query 30 RRKMAASAGASSLAAQAEAVYRERQQIEEKMEALASFLTAEGMPGLKGPLVDPEGFPRAD 89
R+ +S + + + R +++IE +++A L ++ G+ PLVD EG+PR+D
Sbjct 7 RQSGGSSQAGVVTVSDVQELMRRKEEIEAQIKANYDVLESQKGIGMNEPLVDCEGYPRSD 66
Query 90 IDVHAVLQARQQLACLKTDHREVQQRLEETLLLLHA 125
+D++ V AR + CL+ DH+ V +++EE L LHA
Sbjct 67 VDLYQVRTARHNIICLQNDHKAVMKQVEEALHQLHA 102
> At5g57950
Length=275
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 39/113 (34%), Positives = 63/113 (55%), Gaps = 8/113 (7%)
Query 28 RPRRKMAASAGASSLAAQAEAVYRERQQIEEKMEALASFLTAEGMPGLKGPLVDPEGFPR 87
R R+ GA+ L A+ A+ +R +E +M ++ L G PGL G L+D EGFPR
Sbjct 22 RTWRREEKMVGAN-LKAETMALMDKRTAMETEMNSIVERLCNPGGPGLSGNLIDSEGFPR 80
Query 88 ADIDVHAVLQARQQLACLKTDHREVQQRLEETLLLLH-------AANAKDKPP 133
DID+ V R++LA L+++H E+ +++ + +LH A++ KD P
Sbjct 81 EDIDIPMVRTERRRLAELRSEHGEITEKINVNIQILHSVRPTSRASSTKDSGP 133
> SPAC2H10.02c
Length=213
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 45/69 (65%), Gaps = 1/69 (1%)
Query 52 ERQQIEEKMEALASFLTAEGMPGLKGPLVDPEGFPRADIDVHAVLQARQQLACLKTDHRE 111
++++IE ++ L L E + + PL+ +GFPR+DIDV ++ AR ++ L+ DHRE
Sbjct 10 KKREIENRLNELEGVLLKERVT-MDTPLLTEDGFPRSDIDVPSIRTARHEIITLRNDHRE 68
Query 112 VQQRLEETL 120
++ ++++ L
Sbjct 69 LEDQIKKVL 77
> YIL007c
Length=220
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 42/79 (53%), Gaps = 2/79 (2%)
Query 53 RQQIEEKMEALASFLTAEGMPGLKGPLVDPEGFPRADIDVHAVLQARQQLACLKTDHREV 112
+ IE ++EA S L +G+ G+ LV P+G+PR+D+DV V R+ + LK D +
Sbjct 38 KTDIETQLEAYFSVLEQQGI-GMDSALVTPDGYPRSDVDVLQVTMIRKNVNMLKNDLNHL 96
Query 113 QQRLEETLLLLHAANAKDK 131
QR LL H N K
Sbjct 97 LQR-SHVLLNQHFDNMNVK 114
> CE25810
Length=197
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 38/65 (58%), Gaps = 1/65 (1%)
Query 44 AQAEAVYRERQQIEEKMEALASFLTAEGMPGLKGPLVDPEGFPRADIDVHAVLQARQQLA 103
++A+ + ++R +++ K++ L L + PL+D EG+P IDV+AV AR L
Sbjct 5 SKAKELLQQRDELDGKIKELMLVLETNN-STMDSPLLDAEGYPLNTIDVYAVRHARHDLI 63
Query 104 CLKTD 108
CL+ D
Sbjct 64 CLRND 68
> Hs20538502
Length=1359
Score = 30.4 bits (67), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 79 LVDPEGFPRADIDVHAVLQARQQLACLKTDHREVQQRLEETLLLLHAANAK 129
V+ E P+ ++V + ++ QLACLK H ++ E LLL+ + K
Sbjct 801 FVEKEPAPQVSLNVKGIKESVLQLACLKYPHLITKEPFEHESLLLNRKDHK 851
> Hs5174501
Length=519
Score = 29.3 bits (64), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 11/34 (32%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 50 YRERQQIEEKMEALASFLTAEGMPGLKGPLVDPE 83
Y++R +EE E ++L + G+PG P++ E
Sbjct 210 YKQRSSLEEHKERCHNYLESMGLPGTLYPVIKEE 243
> At5g14580
Length=991
Score = 28.5 bits (62), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 12/41 (29%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 28 RPRRKMAASAGASSLAAQAEAVYRERQQIEEKMEALASFLT 68
+P+RK A+ AG + ++ VY E + E + ++ S +T
Sbjct 751 KPKRKPASDAGKDPVMKESSTVYIENSSVGEIVASMPSIVT 791
> 7298144
Length=3319
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 14/55 (25%), Positives = 28/55 (50%), Gaps = 1/55 (1%)
Query 39 ASSLAAQAEAVYRERQQIEEKMEALASFLTAEGMPGLKGPLVDPEGFPRADIDVH 93
S A A+Y++ + + ++ F T+E + G+ + DP GFP +++H
Sbjct 3136 GSYFPGDAYAIYKKNFNVGKYLDLETEFRTSE-LSGILLSVSDPNGFPALSLELH 3189
> CE00246
Length=1744
Score = 27.3 bits (59), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 18/29 (62%), Gaps = 1/29 (3%)
Query 58 EKMEALASFLTAEGMPGLKGPLVDPEGFP 86
+ M L A+G+PGL GP V P+G+P
Sbjct 721 DGMPGLPGDRGADGLPGLPGP-VGPDGYP 748
Lambda K H
0.316 0.127 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1461889840
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40