bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2669_orf1
Length=574
Score E
Sequences producing significant alignments: (Bits) Value
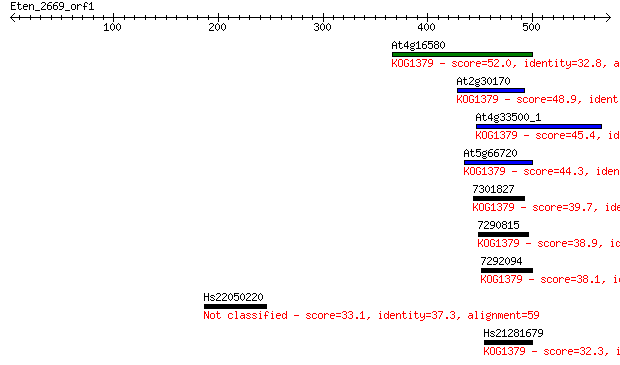
At4g16580 52.0 4e-06
At2g30170 48.9 3e-05
At4g33500_1 45.4 3e-04
At5g66720 44.3 8e-04
7301827 39.7 0.016
7290815 38.9 0.035
7292094 38.1 0.058
Hs22050220 33.1 1.9
Hs21281679 32.3 3.0
> At4g16580
Length=335
Score = 52.0 bits (123), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 44/144 (30%), Positives = 68/144 (47%), Gaps = 15/144 (10%)
Query 366 NHKHSTSGGGCHRGAPRYVAMSDCLGEYPEILVQLLRGPLAKR-----HPEVDLLRLQRE 420
N K + +RGA R++ ++ + + + L L+ R P+V L +
Sbjct 9 NEKSTICAYFAYRGAKRWIYLNQ---QRRGMGFRGLHSSLSNRLSAGNAPDVSLDNSVTD 65
Query 421 AKPKQDSEEPARTDLC-----LWIGAFSIPRDDKRYRGGEDAWFLDASNNAFGVADGVSE 475
+ + DS + LC L G+ +P DK GGEDA F+ A A GVADGV
Sbjct 66 EQVR-DSSDSVAAKLCTKPLKLVSGSCYLPHPDKEATGGEDAHFICAEEQALGVADGVGG 124
Query 476 WEDLAGINPQEYAQDLMQGTQEAI 499
W +L GI+ Y+++LM + AI
Sbjct 125 WAEL-GIDAGYYSRELMSNSVNAI 147
> At2g30170
Length=283
Score = 48.9 bits (115), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 40/65 (61%), Gaps = 2/65 (3%)
Query 429 EPARTDLCLWIGAFSIPRDDKRYRGGEDAWFLDASN-NAFGVADGVSEWEDLAGINPQEY 487
+P R +L L +G +IP DK +GGEDA+F+ + VADGVS W + ++P +
Sbjct 39 QPLRPELSLSVGIHAIPHPDKVEKGGEDAFFVSSYRGGVMAVADGVSGWAE-QDVDPSLF 97
Query 488 AQDLM 492
+++LM
Sbjct 98 SKELM 102
> At4g33500_1
Length=702
Score = 45.4 bits (106), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 35/119 (29%), Positives = 55/119 (46%), Gaps = 32/119 (26%)
Query 447 DDKRYRGGEDAWFLDASNNAFGVADGVSEWEDLAGINPQEYAQDLMQGTQEAIGKIQTEQ 506
D + G EDA+F+ + +N G+ADGVS+W GIN YAQ+LM ++ I
Sbjct 474 DVQALAGREDAYFI-SHHNWIGIADGVSQW-SFEGINKGMYAQELMSNCEKIISN----- 526
Query 507 RSRTKQTETEGTGGGIEADMQTGRQTCNPADVAKEALTRAYHKARNFGSSTAIVGVLDG 565
+T + +P V L R+ ++ ++ GSSTA++ LD
Sbjct 527 --------------------ETAK-ISDPVQV----LHRSVNETKSSGSSTALIAHLDN 560
> At5g66720
Length=414
Score = 44.3 bits (103), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 37/65 (56%), Gaps = 1/65 (1%)
Query 435 LCLWIGAFSIPRDDKRYRGGEDAWFLDASNNAFGVADGVSEWEDLAGINPQEYAQDLMQG 494
L L G+ +P +K GGEDA F+ A GVADGV W ++ G+N ++++LM
Sbjct 168 LRLVSGSCYLPHPEKEATGGEDAHFICDEEQAIGVADGVGGWAEV-GVNAGLFSRELMSY 226
Query 495 TQEAI 499
+ AI
Sbjct 227 SVSAI 231
> 7301827
Length=314
Score = 39.7 bits (91), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 30/51 (58%), Gaps = 3/51 (5%)
Query 444 IPRDDKRYRGGEDAWFLDAS--NNAFGVADGVSEWEDLAGINPQEYAQDLM 492
P + R GED+WF+ ++ GVADGV W DL G++ +A++LM
Sbjct 58 FPGERSNQRFGEDSWFVSSTPLAEVMGVADGVGGWRDL-GVDAGRFAKELM 107
> 7290815
Length=374
Score = 38.9 bits (89), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 31/50 (62%), Gaps = 3/50 (6%)
Query 449 KRYRGGEDAWFLDASNNA--FGVADGVSEWEDLAGINPQEYAQDLMQGTQ 496
R + GEDAWF+ +S A GVADGV W + G++P +++ LM+ +
Sbjct 127 NRGKFGEDAWFMSSSPQACIMGVADGVGGWRNY-GVDPGKFSMTLMRSCE 175
> 7292094
Length=321
Score = 38.1 bits (87), Expect = 0.058, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 29/50 (58%), Gaps = 3/50 (6%)
Query 452 RGGEDAWFL--DASNNAFGVADGVSEWEDLAGINPQEYAQDLMQGTQEAI 499
+ GED+WF AS + GVADGV W GI+P E++ LM+ + +
Sbjct 76 KYGEDSWFKASTASADVMGVADGVGGWRSY-GIDPGEFSSFLMRTCERLV 124
> Hs22050220
Length=161
Score = 33.1 bits (74), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 31/68 (45%), Gaps = 9/68 (13%)
Query 187 EGRQSQWSEGGLHGHLRQPHHRLPPTASPSSNV---------RGRREERGPPAEQTEAAP 237
EG + W G G+ + HR P SP V RG ++ GP AE+ E AP
Sbjct 18 EGVKEPWGSGNNQGNYLEFEHRRDPYPSPRDTVWATCCLRGHRGGKQLLGPQAEEEETAP 77
Query 238 AAGWWIRS 245
G ++R+
Sbjct 78 QMGRFMRT 85
> Hs21281679
Length=304
Score = 32.3 bits (72), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 28/48 (58%), Gaps = 3/48 (6%)
Query 454 GEDAWFL--DASNNAFGVADGVSEWEDLAGINPQEYAQDLMQGTQEAI 499
G+DA F+ S + GVADGV W D G++P +++ LM+ + +
Sbjct 59 GDDACFVARHRSADVLGVADGVGGWRDY-GVDPSQFSGTLMRTCERLV 105
Lambda K H
0.314 0.132 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 15928698760
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40