bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2663_orf1
Length=137
Score E
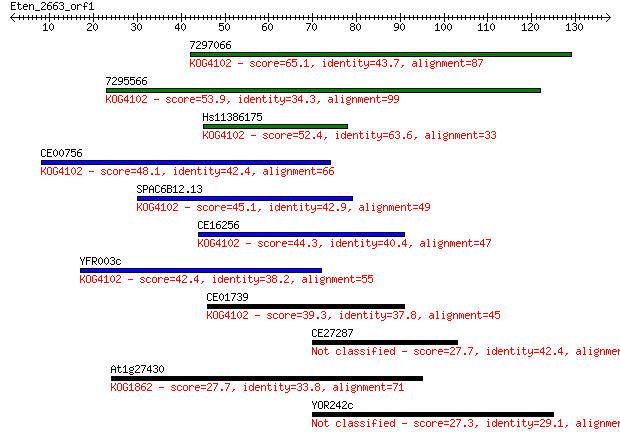
Sequences producing significant alignments: (Bits) Value
7297066 65.1 4e-11
7295566 53.9 9e-08
Hs11386175 52.4 3e-07
CE00756 48.1 4e-06
SPAC6B12.13 45.1 4e-05
CE16256 44.3 7e-05
YFR003c 42.4 3e-04
CE01739 39.3 0.002
CE27287 27.7 6.9
At1g27430 27.7 7.4
YOR242c 27.3 8.3
> 7297066
Length=163
Score = 65.1 bits (157), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 38/90 (42%), Positives = 52/90 (57%), Gaps = 12/90 (13%)
Query 42 EHPK---QVAWVEGTLDNEDLGRKSSKVCCIYHKPVAFGESSSSSSSSSEDELDSNGNVR 98
EHP+ +VA+ G +DNE L RK SK CCIY KP+AFGE SSSED+ D
Sbjct 43 EHPRNERRVAFHAGIIDNEHLNRKKSKCCCIYKKPLAFGE------SSSEDDEDCEHCFG 96
Query 99 RVKKKKRAKRSRREEHVWGDKHCSQGAADE 128
+K+ +R+ + H GDK C++ + E
Sbjct 97 HPEKR---QRNAKHNHNHGDKPCTEASHPE 123
> 7295566
Length=228
Score = 53.9 bits (128), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 34/99 (34%), Positives = 50/99 (50%), Gaps = 4/99 (4%)
Query 23 IVPPAASTAATPQEQQETTEHPKQVAWVEGTLDNEDLGRKSSKVCCIYHKPVAFGESSSS 82
+ P AA T + Q+ + V + G +DNE + R+ SK CCIY KP FGE
Sbjct 49 LQPVAAPTMYCLRLAQQRPITERHVHFHAGVIDNEHMNRRKSKCCCIYRKPHPFGE---- 104
Query 83 SSSSSEDELDSNGNVRRVKKKKRAKRSRREEHVWGDKHC 121
SSSS++DE + V+ + R ++ R +E G C
Sbjct 105 SSSSTDDECEHCFGHPEVRTRNRLEKQRIQEQQNGCSCC 143
> Hs11386175
Length=126
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 21/33 (63%), Positives = 25/33 (75%), Gaps = 0/33 (0%)
Query 45 KQVAWVEGTLDNEDLGRKSSKVCCIYHKPVAFG 77
K+V W T+DNE +GR+SSK CCIY KP AFG
Sbjct 39 KKVEWTSDTVDNEHMGRRSSKCCCIYEKPRAFG 71
> CE00756
Length=107
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 36/70 (51%), Gaps = 10/70 (14%)
Query 8 ASQTRTETITPAVIHIVPPA----ASTAATPQEQQETTEHPKQVAWVEGTLDNEDLGRKS 63
AS T T T+T VPP+ +P Q +T + V W T+DNE +G+K
Sbjct 9 ASSTETSTVT------VPPSREQNLVLHLSPNPAQPSTSERRHVVWATETVDNEGMGKKK 62
Query 64 SKVCCIYHKP 73
SK CCIY KP
Sbjct 63 SKCCCIYKKP 72
> SPAC6B12.13
Length=104
Score = 45.1 bits (105), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 30/54 (55%), Gaps = 5/54 (9%)
Query 30 TAATPQEQQETTEHP-----KQVAWVEGTLDNEDLGRKSSKVCCIYHKPVAFGE 78
+A+ E+ E H ++V W T+DNE + +K SKVCCI+HK F E
Sbjct 24 SASISHEESENVLHLQPEPVRRVRWTVSTVDNEHMNKKKSKVCCIFHKQRKFDE 77
> CE16256
Length=132
Score = 44.3 bits (103), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 44 PKQVAWVEGTLDNEDLGRKSSKVCCIYHKPVAFGESSSSSSSSSEDE 90
P V W EG +DNE +GR S CCIY P + + S+ + E E
Sbjct 35 PPHVTWAEGVVDNEHMGRLKSNCCCIYVAPRQWDDPSTWEPNEYETE 81
> YFR003c
Length=155
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 30/58 (51%), Gaps = 11/58 (18%)
Query 17 TPAVIHIVP---PAASTAATPQEQQETTEHPKQVAWVEGTLDNEDLGRKSSKVCCIYH 71
PAV+ + P S A P T H V W E +DNE++ +K +K+CCI+H
Sbjct 26 VPAVLQLRATQDPPRSQEAMP------TRH--NVRWEENVIDNENMNKKKTKICCIFH 75
> CE01739
Length=109
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 23/45 (51%), Gaps = 0/45 (0%)
Query 46 QVAWVEGTLDNEDLGRKSSKVCCIYHKPVAFGESSSSSSSSSEDE 90
+V W G +DNE +GR S CCIY P + + S+ E E
Sbjct 31 RVTWGAGVIDNEHMGRLKSNCCCIYTPPRVWDDPSTWEPEEHETE 75
> CE27287
Length=691
Score = 27.7 bits (60), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 18/33 (54%), Gaps = 0/33 (0%)
Query 70 YHKPVAFGESSSSSSSSSEDELDSNGNVRRVKK 102
YHKPV F S + SEDEL + + R V +
Sbjct 377 YHKPVRFAVRSLGWTDISEDELTAEKSSRAVNR 409
> At1g27430
Length=1453
Score = 27.7 bits (60), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 33/79 (41%), Gaps = 9/79 (11%)
Query 24 VPPAASTAATPQEQQETTEHPKQVAW--VEGT------LDNEDLGRKSSKVCCIYHKPVA 75
+P +TAAT + Q E HP V W +EG L N G C I ++P+
Sbjct 581 IPVEVTTAAT-RNQNENKLHPFGVLWSELEGGSTPVNPLPNRSSGAMGEPSCSIENRPIN 639
Query 76 FGESSSSSSSSSEDELDSN 94
+S + S D L N
Sbjct 640 SRRNSQIDPNISLDALSGN 658
> YOR242c
Length=371
Score = 27.3 bits (59), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 16/55 (29%), Positives = 27/55 (49%), Gaps = 5/55 (9%)
Query 70 YHKPVAFGESSSSSSSSSEDELDSNGNVRRVKKKKRAKRSRREEHVWGDKHCSQG 124
+ K FG S + S + +N N RR+KK+ + ++ +W +H SQG
Sbjct 54 FAKAYLFGREIGSCGTDSYTPVGANVNKRRLKKE-----DKNDQQLWKRQHHSQG 103
Lambda K H
0.306 0.120 0.342
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1461889840
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40