bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
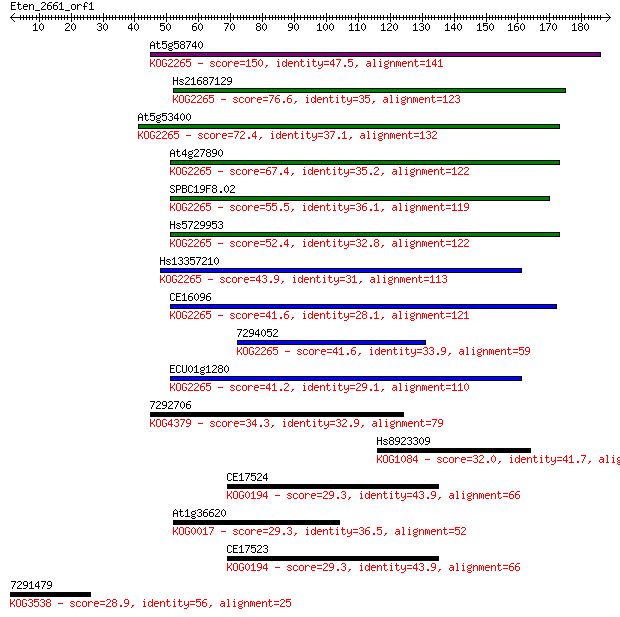
Query= Eten_2661_orf1
Length=188
Score E
Sequences producing significant alignments: (Bits) Value
At5g58740 150 1e-36
Hs21687129 76.6 2e-14
At5g53400 72.4 4e-13
At4g27890 67.4 2e-11
SPBC19F8.02 55.5 6e-08
Hs5729953 52.4 5e-07
Hs13357210 43.9 2e-04
CE16096 41.6 8e-04
7294052 41.6 0.001
ECU01g1280 41.2 0.001
7292706 34.3 0.13
Hs8923309 32.0 0.82
CE17524 29.3 4.6
At1g36620 29.3 4.9
CE17523 29.3 5.3
7291479 28.9 5.9
> At5g58740
Length=158
Score = 150 bits (379), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 67/141 (47%), Positives = 93/141 (65%), Gaps = 0/141 (0%)
Query 45 HDGRLLYEWEQDLTDVHIYLKPPPKTEAKEISVRISPNKLAVGLIGKPPLFDEELSSTVE 104
H+G+ ++EW+Q L +V++Y+ PP K +I + VG+ G PP + +LS+ V+
Sbjct 15 HNGQKVFEWDQTLEEVNMYITLPPNVHPKSFHCKIQSKHIEVGIKGNPPYLNHDLSAPVK 74
Query 105 TAASFWMMEDGELHIQLGKMRKGEVWKSALKGHATLNPLAAEEVQKKLMLERFGEEHPGF 164
T SFW +ED +HI L K KG+ W S + G L+P A + QK+LML+RF EE+PGF
Sbjct 75 TDCSFWTLEDDIMHITLQKREKGQTWASPILGQGQLDPYATDLEQKRLMLQRFQEENPGF 134
Query 165 DFSGATFSGSAPDPRSFMGGI 185
DFS A FSG+ PDPRSFMGGI
Sbjct 135 DFSQAQFSGNCPDPRSFMGGI 155
> Hs21687129
Length=157
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 43/126 (34%), Positives = 71/126 (56%), Gaps = 4/126 (3%)
Query 52 EWEQDLTDVHIYLKPPPKTEAKEISVRISPNKLAVGLIGKPPLFDEELSSTVETAASFWM 111
+W Q L +V I ++ PP T A++I + +A+ + G+ L + ST+ + W
Sbjct 20 QWYQTLEEVFIEVQVPPGTRAQDIQCGLQSRHVALSVGGREILKGKLFDSTIADEGT-WT 78
Query 112 MEDGEL-HIQLGKMRK--GEVWKSALKGHATLNPLAAEEVQKKLMLERFGEEHPGFDFSG 168
+ED ++ I L K ++ W S L+ +P +++Q+KL LERF +E+PGFDFSG
Sbjct 79 LEDRKMVRIVLTKTKRDAANCWTSLLESEYAADPWVQDQMQRKLTLERFQKENPGFDFSG 138
Query 169 ATFSGS 174
A SG+
Sbjct 139 AEISGN 144
> At5g53400
Length=304
Score = 72.4 bits (176), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 49/170 (28%), Positives = 78/170 (45%), Gaps = 38/170 (22%)
Query 41 VPVRHDGRLL--YEWEQDLTDVHIYLKPPPKTEAKEISVRISPNKLAVGLIGKPPLFDEE 98
VP + +G L Y W Q+L +V + + P T+A+ + I N+L VGL G+ P+ D E
Sbjct 135 VPNKGNGTDLENYSWIQNLQEVTVNIPVPTGTKARTVVCEIKKNRLKVGLKGQDPIVDGE 194
Query 99 LSSTVETAASFWMMEDGE-LHIQLGKMRKGEVWKSALKGHATLNPL-------------- 143
L +V+ +W +ED + + I L K + E WK +KG ++
Sbjct 195 LYRSVKPDDCYWNIEDQKVISILLTKSDQMEWWKCCVKGEPEIDTQKVEPETSKLGDLDP 254
Query 144 ---------------------AAEEVQKKLMLERFGEEHPGFDFSGATFS 172
+EE+QK+ +L++F EHP DFS A F+
Sbjct 255 ETRSTVEKMMFDQRQKQMGLPTSEELQKQEILKKFMSEHPEMDFSNAKFN 304
> At4g27890
Length=293
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 43/158 (27%), Positives = 72/158 (45%), Gaps = 36/158 (22%)
Query 51 YEWEQDLTDVHIYLKPPPKTEAKEISVRISPNKLAVGLIGKPPLFDEELSSTVETAASFW 110
Y W Q+L +V I + P T+++ ++ I N+L VGL G+ + D E ++V+ FW
Sbjct 136 YSWGQNLQEVTINIPMPEGTKSRSVTCEIKKNRLKVGLKGQDLIVDGEFFNSVKPDDCFW 195
Query 111 MMEDGEL-HIQLGKMRKGEVWKSALKGHATLNPLAAE----------------------- 146
+ED ++ + L K + E WK +KG ++ E
Sbjct 196 NIEDQKMISVLLTKQDQMEWWKYCVKGEPEIDTQKVEPETSKLGDLDPETRASVEKMMFD 255
Query 147 ------------EVQKKLMLERFGEEHPGFDFSGATFS 172
E++KK ML++F ++PG DFS A F+
Sbjct 256 QRQKQMGLPRSDEIEKKDMLKKFMAQNPGMDFSNAKFN 293
> SPBC19F8.02
Length=166
Score = 55.5 bits (132), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 43/152 (28%), Positives = 64/152 (42%), Gaps = 33/152 (21%)
Query 51 YEWEQDLTDVHIYLKPPPKTEAKEISVRISPN--KLAVGLIGKPPLFDEELSSTVETAAS 108
YEW+Q + DV I + P T AK + V +S + K+ + + + L L + S
Sbjct 11 YEWDQTIADVDIVIHVPKGTRAKSLQVDMSNHDLKIQINVPERKVLLSGPLEKQINLDES 70
Query 109 FWMMEDGE-LHIQLGKMRKGEVWKSALKGH-----------------------ATLNPLA 144
W +E+ E L I L K K E W +KGH AT+ +
Sbjct 71 TWTVEEQERLVIHLEKSNKMEWWSCVIKGHPSIDIGSIEPENSKLSDLDEETRATVEKMM 130
Query 145 AEEVQK-------KLMLERFGEEHPGFDFSGA 169
E+ QK K +L+ F ++HP DFS
Sbjct 131 LEQSQKRTDEQKRKDVLQNFMKQHPELDFSNV 162
> Hs5729953
Length=331
Score = 52.4 bits (124), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 40/160 (25%), Positives = 69/160 (43%), Gaps = 38/160 (23%)
Query 51 YEWEQDLT--DVHIYLKPPPKTEAKEISVRISPNKLAVGLIGKPPLFDEELSSTVETAAS 108
Y W Q L+ D+ + + + K++ V I L VGL G+P + D EL + V+ S
Sbjct 172 YRWTQTLSELDLAVPFCVNFRLKGKDMVVDIQRRHLRVGLKGQPAIIDGELYNEVKVEES 231
Query 109 FWMMEDGE-LHIQLGKMRKGEVWKSALKGHATLNPL------------------------ 143
W++EDG+ + + L K+ K E W + +N
Sbjct 232 SWLIEDGKVVTVHLEKINKMEWWSRLVSSDPEINTKKINPENSKLSDLDSETRSMVEKMM 291
Query 144 -----------AAEEVQKKLMLERFGEEHPGFDFSGATFS 172
++E +K+ +L++F ++HP DFS A F+
Sbjct 292 YDQRQKSMGLPTSDEQKKQEILKKFMDQHPEMDFSKAKFN 331
> Hs13357210
Length=220
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 35/120 (29%), Positives = 65/120 (54%), Gaps = 13/120 (10%)
Query 48 RLLYEWEQDLTDVHIYLKPPPK--TEAKEISVRISPNKLAVGLI---GKPPLFDEELSST 102
R Y W QD TD+ + + P PK + K++SV +S + + V ++ G+ L + +L+
Sbjct 46 RENYTWSQDYTDLEVRV-PVPKHVVKGKQVSVALSSSSIRVAMLEENGERVLMEGKLTHK 104
Query 103 VETAASFWMMEDGE-LHIQLGKMRKGEVWKSA-LKGHATLNPLAAEEVQKKLMLERFGEE 160
+ T +S W +E G+ + + L K+ GE W +A L+G P+ +++ K+ + EE
Sbjct 105 INTESSLWSLEPGKCVLVNLSKV--GEYWWNAILEGE---EPIDIDKINKERSMATVDEE 159
> CE16096
Length=320
Score = 41.6 bits (96), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 34/159 (21%), Positives = 69/159 (43%), Gaps = 38/159 (23%)
Query 51 YEWEQDLTDVHIYLKPPP--KTEAKEISVRISPNKLAVGLIGKPPLFDEELSSTVETAAS 108
Y+W Q L ++ + + +++++ V+I ++VGL + P+ D +L ++
Sbjct 161 YQWTQTLQELEVKIPIAAGFAIKSRDVVVKIEKTSVSVGLKNQAPIVDGKLPHAIKVENC 220
Query 109 FWMMEDGE-LHIQLGKMRKGEVWK---------------------SALKGH--ATLNPLA 144
W++E+G+ + + L K+ E W S L G A + +
Sbjct 221 NWVIENGKAIVLTLEKINDMEWWNRFLDSDPPINTKEVKPENSKLSDLDGETRAMVEKMM 280
Query 145 AEEVQKKL------------MLERFGEEHPGFDFSGATF 171
++ QK++ ML++F ++HP DFS A
Sbjct 281 YDQRQKEMGLPTSDEKKKHDMLQQFMKQHPEMDFSNAKI 319
> 7294052
Length=332
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 34/60 (56%), Gaps = 1/60 (1%)
Query 72 AKEISVRISPNKLAVGLIGKPPLFDEELSSTVETAASFWMMEDGE-LHIQLGKMRKGEVW 130
A+++ + I L VG+ G+ P+ D EL V+T S W+++D + + I L K+ K W
Sbjct 212 ARDLVISIGKKSLKVGIKGQTPIIDGELCGEVKTEESVWVLQDSKTVMITLDKINKMNWW 271
> ECU01g1280
Length=131
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 32/125 (25%), Positives = 60/125 (48%), Gaps = 17/125 (13%)
Query 51 YEWEQDLTDVHIYLKPPPKTEAKEISVRISPNKLAVGLIGKPPLFDEELSSTVETAASFW 110
Y W+Q+L +++I ++ I +RI K+ V G+ + D EL V+ ++ +W
Sbjct 7 YTWDQELNEINIQFPVTGDADSSAIKIRIVGKKICVKNQGE-IVIDGELLHEVDVSSLWW 65
Query 111 MMEDGELHIQLGKMRKGEVWKSALKG-------------HATLNPLAAE--EVQKKLMLE 155
++ + + + K ++ E W S L G HA ++ L AE EV +K+M
Sbjct 66 VINGDVVDVNVTK-KRNEWWDSLLVGSESVDVQKLAENKHADMSMLDAEAREVVEKMMHN 124
Query 156 RFGEE 160
G++
Sbjct 125 TSGKD 129
> 7292706
Length=580
Score = 34.3 bits (77), Expect = 0.13, Method: Composition-based stats.
Identities = 26/85 (30%), Positives = 39/85 (45%), Gaps = 7/85 (8%)
Query 45 HDGRL------LYEWEQDLTDVHIYLKPPPKTEAKEISVRISPNKLAVGLIGKPPLFDEE 98
HDG+L Y W Q +V I P AKE +V S + V + + +FDEE
Sbjct 284 HDGQLENGGPPSYSWSQTDNNVLIRFNVPSTASAKEFNVSGSKTTVVVKHLDE-VIFDEE 342
Query 99 LSSTVETAASFWMMEDGELHIQLGK 123
L ++V S + + + L + L K
Sbjct 343 LYASVGDDLSAFKLAENGLELSLTK 367
> Hs8923309
Length=706
Score = 32.0 bits (71), Expect = 0.82, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 26/56 (46%), Gaps = 8/56 (14%)
Query 116 ELHIQLGK------MRKGEVWKSALKG--HATLNPLAAEEVQKKLMLERFGEEHPG 163
EL Q+ K + K +W SAL + NP A EV + + FG EHPG
Sbjct 352 ELGFQIKKKTKRLYINKANIWNSALDAFRNRNFNPSYAIEVAYVIENDNFGSEHPG 407
> CE17524
Length=262
Score = 29.3 bits (64), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 29/87 (33%), Positives = 37/87 (42%), Gaps = 22/87 (25%)
Query 69 KTEAKEISVRISPNKLAVGLIGKPPLFDEELSSTVETAASF------------------- 109
KT A VR S +K+ V IG P + E ++ V T SF
Sbjct 55 KTSANHFVVRESDHKVYVDKIGFPTII-ELVNHYVSTKESFSTKDGTVKAVLKQAVERQK 113
Query 110 WMM--EDGELHIQLGKMRKGEVWKSAL 134
W + ED EL +LG+ GEVWK L
Sbjct 114 WELYHEDVELTKKLGEGAFGEVWKGKL 140
> At1g36620
Length=1152
Score = 29.3 bits (64), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 29/63 (46%), Gaps = 11/63 (17%)
Query 52 EWEQDLTDVH-----------IYLKPPPKTEAKEISVRISPNKLAVGLIGKPPLFDEELS 100
+WE DVH IY+KPPP + + S+ K GL P + E+LS
Sbjct 1036 DWELHQMDVHNAFLHGDLKEDIYMKPPPGFKTTDPSLVCKLKKSIYGLKQAPRCWFEKLS 1095
Query 101 STV 103
+++
Sbjct 1096 TSL 1098
> CE17523
Length=430
Score = 29.3 bits (64), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 29/87 (33%), Positives = 37/87 (42%), Gaps = 22/87 (25%)
Query 69 KTEAKEISVRISPNKLAVGLIGKPPLFDEELSSTVETAASF------------------- 109
KT A VR S +K+ V IG P + E ++ V T SF
Sbjct 55 KTSANHFVVRESDHKVYVDKIGFPTII-ELVNHYVSTKESFSTKDGTVKAVLKQAVERQK 113
Query 110 WMM--EDGELHIQLGKMRKGEVWKSAL 134
W + ED EL +LG+ GEVWK L
Sbjct 114 WELYHEDVELTKKLGEGAFGEVWKGKL 140
> 7291479
Length=1091
Score = 28.9 bits (63), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 16/30 (53%), Gaps = 5/30 (16%)
Query 1 FVKFREY-----NSSFKPNTRSSPAGKLDC 25
F+K+R Y N F P TR SP GK C
Sbjct 986 FIKYRYYLVNGRNGHFPPGTRCSPVGKRYC 1015
Lambda K H
0.316 0.134 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3094507556
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40