bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2646_orf1
Length=315
Score E
Sequences producing significant alignments: (Bits) Value
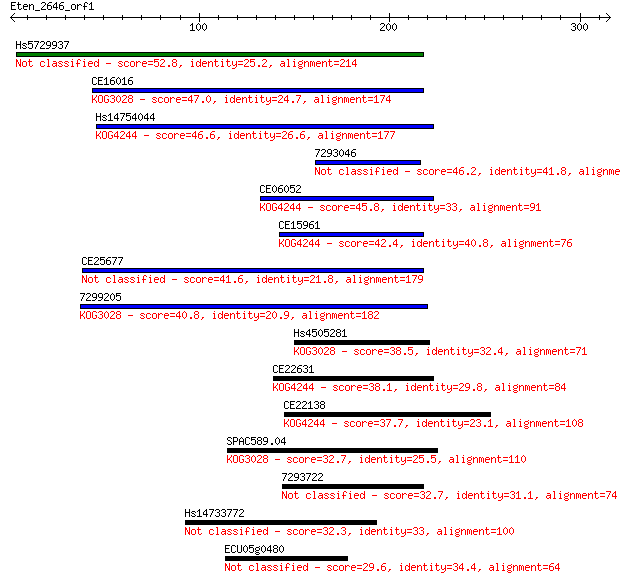
Hs5729937 52.8 1e-06
CE16016 47.0 5e-05
Hs14754044 46.6 6e-05
7293046 46.2 1e-04
CE06052 45.8 1e-04
CE15961 42.4 0.001
CE25677 41.6 0.002
7299205 40.8 0.004
Hs4505281 38.5 0.021
CE22631 38.1 0.024
CE22138 37.7 0.028
SPAC589.04 32.7 1.1
7293722 32.7 1.1
Hs14733772 32.3 1.3
ECU05g0480 29.6 9.5
> Hs5729937
Length=263
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 54/214 (25%), Positives = 88/214 (41%), Gaps = 30/214 (14%)
Query 4 AKVPLTSLGTAVNAENRDARGGSLPDDVLCIGNCDAHTCESVEKAIRLAVTELLQQAVQT 63
KVP +G V +E G + V G+ + E V+KA A EL+ + T
Sbjct 70 GKVPFIHVGNQVVSE-----LGPIVQFVKAKGHSLSDGLEEVQKAEMKAYMELVNNMLLT 124
Query 64 ALCFYLWKDEATFRGFTRPLHKNTLGFVYGSYYCWVMKRSACSSRVACRSPCALPEDLKR 123
A + W DEAT T YGS Y W + + ++KR
Sbjct 125 AELYLQWCDEATVGEITHAR--------YGSPYPWPLNHILAYQK---------QWEVKR 167
Query 124 QFPSCVDSDSTHELVTLEELSVIDKLRRALRVVEQLLQGKQYLGGYKASVLDALVFAHLA 183
+ + T + V++ + + + + Q L + Y + + LDALVF HL
Sbjct 168 KMKAIGWGKKTLD-------QVLEDVDQCCQALSQRLGTQPYFFNKQPTELDALVFGHLY 220
Query 184 ILFSLPLPDHRELQAMLLNRGSLLHYCRRVQEVY 217
+ + L + EL + N +LL +CRR+++ Y
Sbjct 221 TILTTQLTND-ELSEKVKNYSNLLAFCRRIEQHY 253
> CE16016
Length=312
Score = 47.0 bits (110), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 43/174 (24%), Positives = 70/174 (40%), Gaps = 30/174 (17%)
Query 44 SVEKAIRLAVTELLQQAVQTALCFYLWKDEATFRGFTRPLHKNTLGFVYGSYYCWVMKRS 103
++EKA A + L + A+ W DE + T+ + + L F Y YY + KR
Sbjct 87 TIEKAQLDAFSCYLHHNLYPAVMHTFWTDELNYNTVTQYWYASHLHFPYNLYY--LEKRR 144
Query 104 ACSSRVACRSPCALPEDLKRQFPSCVDSDSTHELVTLEELSVIDKLRRALRVVEQLLQGK 163
+ R+ L + ++ + AL + L
Sbjct 145 KKALRL---------------------------LAGKNDTEILKEAFMALNTLSTKLGDN 177
Query 164 QYLGGYKASVLDALVFAHLAILFSLPLPDHRELQAMLLNRGSLLHYCRRVQEVY 217
++ G K + LDALVF +LA L +PLP+ R LQ L +L+ + V +Y
Sbjct 178 KFFCGNKPTSLDALVFGYLAPLLRVPLPNDR-LQVQLSACPNLVRFVETVSSIY 230
> Hs14754044
Length=409
Score = 46.6 bits (109), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 47/180 (26%), Positives = 80/180 (44%), Gaps = 32/180 (17%)
Query 46 EKAIRLAVTELLQQAVQTALCFYLWKDEATFRGFTRPLHKNTLGFVYGSYYCWVMKRSAC 105
E+AI AVT+++++ L + W D TR + + G + + WV+
Sbjct 183 ERAISRAVTKMVEEHFYWTLAYCQWVDNLNE---TRKMLSLSGGGPFSNLLRWVV----- 234
Query 106 SSRVACRSPCALPEDL-KRQFPSCVDSDSTHELVTLEELSVIDKLRRALRVVEQLLQGKQ 164
C + + + KR+ H + E + + + +R + LL K+
Sbjct 235 ---------CHITKGIVKREM-------HGHGIGRFSEEEIYMLMEKDMRSLAGLLGDKK 278
Query 165 YLGGYKASVLDALVFAHLA-ILFSLP-LPDHRELQAMLLNRGSLLHYCRRVQEVYRIWPE 222
Y+ G K S LDA VF HLA +++LP R ++ L+N L YC R++ + WPE
Sbjct 279 YIMGPKLSTLDATVFGHLAQAMWTLPGTRPERLIKGELIN---LAMYCERIRR--KFWPE 333
> 7293046
Length=281
Score = 46.2 bits (108), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 23/55 (41%), Positives = 32/55 (58%), Gaps = 1/55 (1%)
Query 161 QGKQYLGGYKASVLDALVFAHLAILFSLPLPDHRELQAMLLNRGSLLHYCRRVQE 215
Q K Y GG + LDALVF H+ + + LP+ EL A+L LL +CRR+ +
Sbjct 203 QAKSYFGGSRPCKLDALVFGHVVAIMTTKLPN-MELAAVLATYPRLLAHCRRIDQ 256
> CE06052
Length=287
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/92 (32%), Positives = 50/92 (54%), Gaps = 6/92 (6%)
Query 132 DSTHELVTLEELSVIDKLRRALRVVEQLLQGKQYLGGYKASVLDALVFAHLAILFSLPLP 191
+ST + E + + L R LRVVE +L K+YL G + +DA VF+ LA+++ P
Sbjct 197 NSTSAIGDFEPAELDELLHRDLRVVETILGNKKYLFGDHIAPVDATVFSQLAVVY---YP 253
Query 192 DHRELQAMLLNR-GSLLHYCRRVQEVYRIWPE 222
+ + +L N +L YC R+++ I+P
Sbjct 254 FYTHISTVLENDFPKILQYCERIRK--EIYPN 283
> CE15961
Length=277
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 31/78 (39%), Positives = 44/78 (56%), Gaps = 4/78 (5%)
Query 142 ELSVIDK-LRRALRVVEQLLQGKQYLGGYKASVLDALVFAHLAILFSLPLPDHRELQAML 200
ELS +D+ L R LR+VE L K++L G + + DA VF+ LA ++ P +H +
Sbjct 196 ELSELDEILHRDLRIVENTLAKKKFLFGEEITAADATVFSQLATVYY-PFRNHIS-DVLE 253
Query 201 LNRGSLLHYCRRV-QEVY 217
+ LL YC RV EVY
Sbjct 254 KDFPKLLEYCERVRHEVY 271
> CE25677
Length=244
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 39/179 (21%), Positives = 78/179 (43%), Gaps = 26/179 (14%)
Query 39 AHTCESVEKAIRLAVTELLQQAVQTALCFYLWKDEATFRGFTRPLHKNTLGFVYGSYYCW 98
+H E+ +R ++ +++ + T F LW + T+ T+ YGS Y W
Sbjct 89 SHLSETQVADMRANIS-MIEHLLTTVEKFVLWNHDETYDKVTK--------LRYGSVYHW 139
Query 99 VMKRSACSSRVACRSPCALPEDLKRQFPSCVDSDSTHELVTLEELSVIDKLRRALRVVEQ 158
SS + + E+L SD + T++E+ ++ + R +
Sbjct 140 -----PLSSVLPFVKRRKILEEL---------SDKDWDTKTMDEVG--EQADKVFRALSA 183
Query 159 LLQGKQYLGGYKASVLDALVFAHLAILFSLPLPDHRELQAMLLNRGSLLHYCRRVQEVY 217
L ++YL G + DAL+F H+ L ++ LP + +L +L+ + +R+++ Y
Sbjct 184 QLGSQKYLTGDLPTEADALLFGHMYTLITVRLP-LTNITNILKKYSNLIEFTKRIEQQY 241
> 7299205
Length=327
Score = 40.8 bits (94), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 38/183 (20%), Positives = 81/183 (44%), Gaps = 20/183 (10%)
Query 38 DAHTCESVEKAIRLAVTELLQQAVQTALCFYLWKDEATFRGFTRPLHKNTLGFVYGSYYC 97
DAH + +K + A + + ++L+ + F TR L+ F + YY
Sbjct 83 DAH-LSTKQKHLSTAYANWVFTNLHAYYHYFLFGEPHNFDTTTRGLYAKRTPFPFNFYYP 141
Query 98 WVMKRSACSSRVACRSPCALPEDLKRQFPSCVDSDSTHELVTLEELSVIDKLRRALRVVE 157
+R AC V + + + L + E ++ ++ + ++
Sbjct 142 SSYQREACDV-VQVMAGFDVNDKLDKH----------------EGDYLVVNAKKVVNLLS 184
Query 158 QLLQGKQYLGGYKASVLDALVFAHLAILFSLPLPDHRELQAMLLNRGSLLHYCRRV-QEV 216
+ L K + G S DA+V+++LAI+F + LP++ LQ + +L+++ R+ +++
Sbjct 185 RKLGRKVWFFGDTYSEFDAIVYSYLAIIFKIALPNN-PLQNHIKGCQNLVNFINRITKDI 243
Query 217 YRI 219
+RI
Sbjct 244 FRI 246
> Hs4505281
Length=317
Score = 38.5 bits (88), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 36/71 (50%), Gaps = 1/71 (1%)
Query 150 RRALRVVEQLLQGKQYLGGYKASVLDALVFAHLAILFSLPLPDHRELQAMLLNRGSLLHY 209
R L ++ Q L +++ G + LDA VF++LA+L LP + LQ L +L Y
Sbjct 175 RECLTLLSQRLGSQKFFFGDAPASLDAFVFSYLALLLQAKLPSGK-LQVHLRGLHNLCAY 233
Query 210 CRRVQEVYRIW 220
C + +Y W
Sbjct 234 CTHILSLYFPW 244
> CE22631
Length=269
Score = 38.1 bits (87), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 41/85 (48%), Gaps = 4/85 (4%)
Query 139 TLEELSVIDKLRRALRVVEQLLQGKQYLGGYKASVLDALVFAHLAILFSLPLPDHRELQA 198
T V+D+ ++ L + L K YL G +D FAHLA L P E++A
Sbjct 171 TFSREEVLDQAKKDLDAISTQLGDKPYLFGSSIKTIDVTAFAHLAELIYTPQFS-PEIRA 229
Query 199 MLLNR-GSLLHYCRRVQEVYRIWPE 222
+ + +++ Y R++E Y WP+
Sbjct 230 YIDEKVPNVMEYVIRIKEKY--WPD 252
> CE22138
Length=321
Score = 37.7 bits (86), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 25/109 (22%), Positives = 50/109 (45%), Gaps = 15/109 (13%)
Query 145 VIDKLRRALRVVEQLLQGKQYLGGYKASVLDALVFAHLAILFSLPL-PDHRELQAMLLNR 203
+I+ L+ + + L K Y G+K + +DA +F+HL ++ P +HR+L +
Sbjct 219 IINIGSEDLKAISKYLGNKHYFHGFKPTKVDACIFSHLCQIYYAPYTSEHRDL--IDGEC 276
Query 204 GSLLHYCRRVQEVYRIWPEGPSFLFGVLSLSDVARGASLRVHSWRQKRI 252
+L Y R++ R +P+ DV S +W+++ +
Sbjct 277 KNLAEYVERIKN--RFYPD----------WDDVTTKFSTDTSNWKKRHV 313
> SPAC589.04
Length=271
Score = 32.7 bits (73), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 28/111 (25%), Positives = 51/111 (45%), Gaps = 8/111 (7%)
Query 115 CALPEDLKRQFPSCVDSDSTHELVTLEELSVIDKLRRALRVVEQLL-QGKQYLGGYKASV 173
LP +KR+ C+ + + TL+ ++++ +A + +LL K + S
Sbjct 164 IGLPSQIKRKI--CLQLNES----TLDFDAILEDASKAFSALSELLGSDKWFFNNESPSF 217
Query 174 LDALVFAHLAILFSLPLPDHRELQAMLLNRGSLLHYCRRVQEVYRIWPEGP 224
LD +FAH I+ LPL + +L+ +L +L RV+ + GP
Sbjct 218 LDVSLFAHAEIINHLPL-KNDQLKVVLGTHKNLTDLTTRVRTLAGYTSAGP 267
> 7293722
Length=269
Score = 32.7 bits (73), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 36/77 (46%), Gaps = 4/77 (5%)
Query 144 SVIDKLRRALRVVEQLLQGKQ---YLGGYKASVLDALVFAHLAILFSLPLPDHRELQAML 200
SVIDK+ + +E L+ + G + LDA+ F HL + + LP+ L +
Sbjct 186 SVIDKVAKCCETLEYKLKESPETPFFYGDQPCELDAIAFGHLFSILTTTLPN-MALAQTV 244
Query 201 LNRGSLLHYCRRVQEVY 217
L+ +CR V E Y
Sbjct 245 QKFQHLVEFCRFVDEKY 261
> Hs14733772
Length=555
Score = 32.3 bits (72), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 33/120 (27%), Positives = 45/120 (37%), Gaps = 23/120 (19%)
Query 93 GSYYCWVMKRSACSSRVACRSPCALPEDLKRQ--FPSCVDSDSTHELVTLEEL--SVIDK 148
S Y MK SAC +V+C + L + K Q P + T L ++ +VI K
Sbjct 206 NSPYSQTMKSSACKIQVSCSNNTHLVSENKEQTTHPELFAGNKTQNLHHMQYFPNNVIPK 265
Query 149 ---LRRALRVVEQLLQGKQYLGGYKASVLDA-------------LVFAHLAILFSLPLPD 192
L R + EQ Q L GYK D L+ H + P+PD
Sbjct 266 QDLLHRCFQEQEQKSQQASVLQGYKNRNQDMSGQQAAQLAQQRYLIHNHANV---FPVPD 322
> ECU05g0480
Length=735
Score = 29.6 bits (65), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 40/67 (59%), Gaps = 7/67 (10%)
Query 114 PCALPEDLKRQFPSCVDSDSTHELVTLEEL--SVIDKLR-RALRVVEQLLQGKQYLGGYK 170
P A+PE ++R+ CVDS +LV ++EL S I +++ AL++VE ++ K+ G
Sbjct 14 PSAMPE-IEREIQRCVDSP---DLVPVKELMNSTIPEVKFYALKIVEHRVRNKKMAGVVP 69
Query 171 ASVLDAL 177
A ++ +
Sbjct 70 AEEIELM 76
Lambda K H
0.322 0.135 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7308161082
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40