bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2594_orf1
Length=170
Score E
Sequences producing significant alignments: (Bits) Value
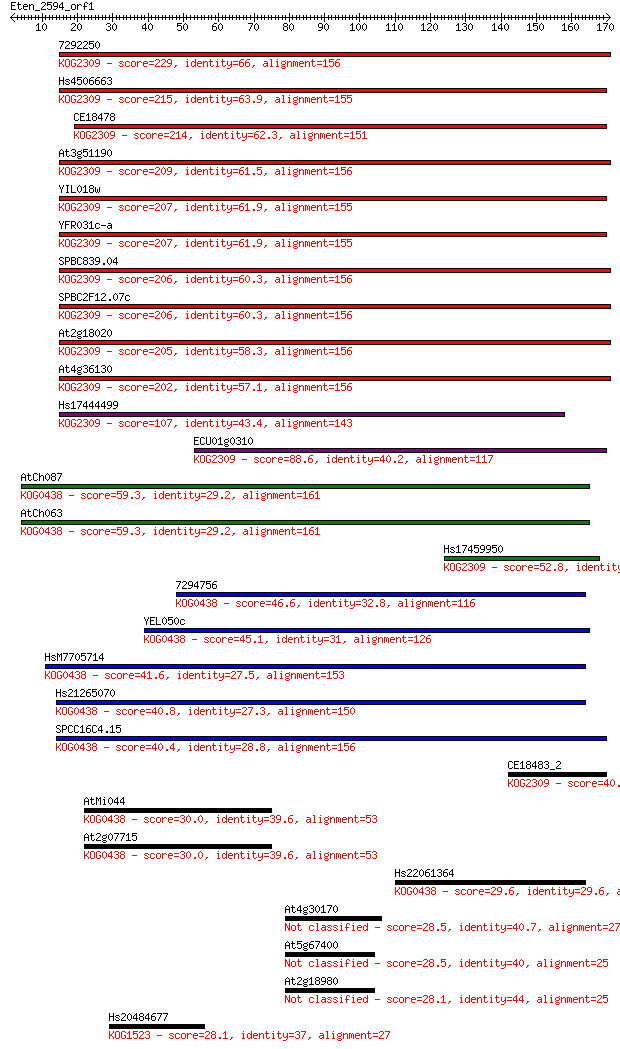
7292250 229 2e-60
Hs4506663 215 3e-56
CE18478 214 1e-55
At3g51190 209 2e-54
YIL018w 207 7e-54
YFR031c-a 207 7e-54
SPBC839.04 206 2e-53
SPBC2F12.07c 206 2e-53
At2g18020 205 4e-53
At4g36130 202 2e-52
Hs17444499 107 9e-24
ECU01g0310 88.6 5e-18
AtCh087 59.3 3e-09
AtCh063 59.3 3e-09
Hs17459950 52.8 3e-07
7294756 46.6 2e-05
YEL050c 45.1 7e-05
HsM7705714 41.6 8e-04
Hs21265070 40.8 0.001
SPCC16C4.15 40.4 0.002
CE18483_2 40.0 0.002
AtMi044 30.0 2.1
At2g07715 30.0 2.5
Hs22061364 29.6 3.0
At4g30170 28.5 6.8
At5g67400 28.5 7.4
At2g18980 28.1 8.3
Hs20484677 28.1 8.6
> 7292250
Length=256
Score = 229 bits (585), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 103/156 (66%), Positives = 134/156 (85%), Gaps = 0/156 (0%)
Query 15 LLLAQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSF 74
++ AQRKG GSVF+AH KRKGAAKLR+LD+AER+GYI+G+V I HDPGRGAPLAVV F
Sbjct 4 VIRAQRKGAGSVFKAHVKKRKGAAKLRSLDFAERSGYIRGVVKDIIHDPGRGAPLAVVHF 63
Query 75 RDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKAGDR 134
RD YRY+I KE +A EG+HTGQF++CG+KA L IGNV+PL ++PEGT++C++EEK GDR
Sbjct 64 RDPYRYKIRKELFIAPEGMHTGQFVYCGRKATLQIGNVMPLSQMPEGTIICNLEEKTGDR 123
Query 135 GTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLP 170
G +A+TSG +ATV+ H+++T KTRV+LPSGA+K +P
Sbjct 124 GRLARTSGNYATVIAHNQDTKKTRVKLPSGAKKVVP 159
> Hs4506663
Length=257
Score = 215 bits (548), Expect = 3e-56, Method: Compositional matrix adjust.
Identities = 99/155 (63%), Positives = 125/155 (80%), Gaps = 0/155 (0%)
Query 15 LLLAQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSF 74
++ QRKG GSVFRAH RKGAA+LRA+D+AER+GYIKG+V I HDPGRGAPLA V F
Sbjct 4 VIRGQRKGAGSVFRAHVKHRKGAARLRAVDFAERHGYIKGIVKDIIHDPGRGAPLAKVVF 63
Query 75 RDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKAGDR 134
RD YR++ E +A EG+HTGQF++CGKKA L IGNVLP+G +PEGT+VC +EEK GDR
Sbjct 64 RDPYRFKKRTELFIAAEGIHTGQFVYCGKKAQLNIGNVLPVGTMPEGTIVCCLEEKPGDR 123
Query 135 GTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTL 169
G +A+ SG +ATV+ H+ ET KTRV+LPSG++K +
Sbjct 124 GKLARASGNYATVISHNPETKKTRVKLPSGSKKVI 158
> CE18478
Length=260
Score = 214 bits (544), Expect = 1e-55, Method: Compositional matrix adjust.
Identities = 94/151 (62%), Positives = 123/151 (81%), Gaps = 0/151 (0%)
Query 19 QRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSFRDAY 78
QRKG G +F++H RKGA+KLR LDYAER+GYIKGLV I HDPGRGAPLA+++FRD Y
Sbjct 8 QRKGAGGIFKSHNKHRKGASKLRPLDYAERHGYIKGLVKDIIHDPGRGAPLAIIAFRDPY 67
Query 79 RYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKAGDRGTIA 138
+Y+ K ++A EG+HTGQFIHCG KA + IGN++P+G +PEGT +C+VE K+GDRG IA
Sbjct 68 KYKTVKTTVVAAEGMHTGQFIHCGAKAQIQIGNIVPVGTLPEGTTICNVENKSGDRGVIA 127
Query 139 KTSGTFATVVGHSEETGKTRVRLPSGARKTL 169
+ SG +ATV+ H+ +T KTR+RLPSGA+K +
Sbjct 128 RASGNYATVIAHNPDTKKTRIRLPSGAKKVV 158
> At3g51190
Length=260
Score = 209 bits (532), Expect = 2e-54, Method: Compositional matrix adjust.
Identities = 96/157 (61%), Positives = 124/157 (78%), Gaps = 1/157 (0%)
Query 15 LLLAQRKGR-GSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAVVS 73
++ AQRKG GSVF++HTH RKG AK R+LDY ERNGY+KGLVT I HDPGRGAPLA V+
Sbjct 4 VIRAQRKGAAGSVFKSHTHHRKGPAKFRSLDYGERNGYLKGLVTEIIHDPGRGAPLARVA 63
Query 74 FRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKAGD 133
FR +RY KE +A EG++TGQ+++CGKKA+L +GNVLPLG +PEG V+C+VE GD
Sbjct 64 FRHPFRYMKQKELFVAAEGMYTGQYLYCGKKANLMVGNVLPLGSIPEGAVICNVELHVGD 123
Query 134 RGTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLP 170
RG +A+ SG +A V+ H+ E+ TRV+LPSG++K LP
Sbjct 124 RGALARASGDYAIVIAHNPESNTTRVKLPSGSKKILP 160
> YIL018w
Length=254
Score = 207 bits (527), Expect = 7e-54, Method: Compositional matrix adjust.
Identities = 96/155 (61%), Positives = 122/155 (78%), Gaps = 0/155 (0%)
Query 15 LLLAQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSF 74
++ QRKG GS+F +HT R+GAAKLR LDYAER+GYI+G+V I HD GRGAPLA V F
Sbjct 4 VIRNQRKGAGSIFTSHTRLRQGAAKLRTLDYAERHGYIRGIVKQIVHDSGRGAPLAKVVF 63
Query 75 RDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKAGDR 134
RD Y+YR+ +E +A EG+HTGQFI+ GKKA L +GNVLPLG VPEGT+V +VEEK GDR
Sbjct 64 RDPYKYRLREEIFIANEGVHTGQFIYAGKKASLNVGNVLPLGSVPEGTIVSNVEEKPGDR 123
Query 135 GTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTL 169
G +A+ SG + ++GH+ + KTRVRLPSGA+K +
Sbjct 124 GALARASGNYVIIIGHNPDENKTRVRLPSGAKKVI 158
> YFR031c-a
Length=254
Score = 207 bits (527), Expect = 7e-54, Method: Compositional matrix adjust.
Identities = 96/155 (61%), Positives = 122/155 (78%), Gaps = 0/155 (0%)
Query 15 LLLAQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSF 74
++ QRKG GS+F +HT R+GAAKLR LDYAER+GYI+G+V I HD GRGAPLA V F
Sbjct 4 VIRNQRKGAGSIFTSHTRLRQGAAKLRTLDYAERHGYIRGIVKQIVHDSGRGAPLAKVVF 63
Query 75 RDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKAGDR 134
RD Y+YR+ +E +A EG+HTGQFI+ GKKA L +GNVLPLG VPEGT+V +VEEK GDR
Sbjct 64 RDPYKYRLREEIFIANEGVHTGQFIYAGKKASLNVGNVLPLGSVPEGTIVSNVEEKPGDR 123
Query 135 GTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTL 169
G +A+ SG + ++GH+ + KTRVRLPSGA+K +
Sbjct 124 GALARASGNYVIIIGHNPDENKTRVRLPSGAKKVI 158
> SPBC839.04
Length=253
Score = 206 bits (524), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 94/156 (60%), Positives = 127/156 (81%), Gaps = 1/156 (0%)
Query 15 LLLAQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSF 74
++ AQRK G +F+AHT RKGAA+LR LD+AER+GYI+G+V I HDPGRGAPLA V+F
Sbjct 4 VIRAQRKS-GGIFQAHTRLRKGAAQLRTLDFAERHGYIRGVVQKIIHDPGRGAPLAKVAF 62
Query 75 RDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKAGDR 134
R+ Y YR + E +A EG++TGQF++CGK A LT+GNVLP+G++PEGT++ +VEEKAGDR
Sbjct 63 RNPYHYRTDVETFVATEGMYTGQFVYCGKNAALTVGNVLPVGEMPEGTIISNVEEKAGDR 122
Query 135 GTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLP 170
G + ++SG + +VGH +TGKTRV+LPSGA+K +P
Sbjct 123 GALGRSSGNYVIIVGHDVDTGKTRVKLPSGAKKVVP 158
> SPBC2F12.07c
Length=253
Score = 206 bits (524), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 94/156 (60%), Positives = 127/156 (81%), Gaps = 1/156 (0%)
Query 15 LLLAQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSF 74
++ AQRK G +F+AHT RKGAA+LR LD+AER+GYI+G+V I HDPGRGAPLA V+F
Sbjct 4 VIRAQRKS-GGIFQAHTRLRKGAAQLRTLDFAERHGYIRGVVQKIIHDPGRGAPLAKVAF 62
Query 75 RDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKAGDR 134
R+ Y YR + E +A EG++TGQF++CGK A LT+GNVLP+G++PEGT++ +VEEKAGDR
Sbjct 63 RNPYHYRTDVETFVATEGMYTGQFVYCGKNAALTVGNVLPVGEMPEGTIISNVEEKAGDR 122
Query 135 GTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLP 170
G + ++SG + +VGH +TGKTRV+LPSGA+K +P
Sbjct 123 GALGRSSGNYVIIVGHDVDTGKTRVKLPSGAKKVVP 158
> At2g18020
Length=258
Score = 205 bits (521), Expect = 4e-53, Method: Compositional matrix adjust.
Identities = 91/156 (58%), Positives = 123/156 (78%), Gaps = 0/156 (0%)
Query 15 LLLAQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSF 74
++ AQRKG GSVF++HTH RKG AK R+LD+ ERNGY+KG+VT I HDPGRGAPLA V+F
Sbjct 4 VIRAQRKGAGSVFKSHTHHRKGPAKFRSLDFGERNGYLKGVVTEIIHDPGRGAPLARVTF 63
Query 75 RDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKAGDR 134
R +R++ KE +A EG++TGQF++CGKKA L +GNVLPL +PEG VVC+VE GDR
Sbjct 64 RHPFRFKKQKELFVAAEGMYTGQFLYCGKKATLVVGNVLPLRSIPEGAVVCNVEHHVGDR 123
Query 135 GTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLP 170
G +A+ SG +A V+ H+ ++ TR++LPSG++K +P
Sbjct 124 GVLARASGDYAIVIAHNPDSDTTRIKLPSGSKKIVP 159
> At4g36130
Length=258
Score = 202 bits (514), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 89/156 (57%), Positives = 121/156 (77%), Gaps = 0/156 (0%)
Query 15 LLLAQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSF 74
++ AQRKG GSVF++HTH RKG AK R+LD+ ERNGY+KG+VT I HDPGRGAPLA V+F
Sbjct 4 VIRAQRKGAGSVFKSHTHHRKGPAKFRSLDFGERNGYLKGVVTEIIHDPGRGAPLARVAF 63
Query 75 RDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKAGDR 134
R +R++ KE +A EG++TGQF++CGKKA L +GNVLPL +PEG V+C+VE GDR
Sbjct 64 RHPFRFKKQKELFVAAEGMYTGQFLYCGKKATLVVGNVLPLRSIPEGAVICNVEHHVGDR 123
Query 135 GTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTLP 170
G A+ SG +A V+ H+ + +R++LPSG++K +P
Sbjct 124 GVFARASGDYAIVIAHNPDNDTSRIKLPSGSKKIVP 159
> Hs17444499
Length=239
Score = 107 bits (268), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 62/143 (43%), Positives = 85/143 (59%), Gaps = 13/143 (9%)
Query 15 LLLAQRKGRGSVFRAHTHKRKGAAKLRALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSF 74
++ QRKG G+ F A + AL + +GY K V H PG GAPLA V F
Sbjct 4 MVCGQRKGAGAAFPAQ--------RACAL-WTAPSGY-KSTVKGAIHYPGCGAPLAKVVF 53
Query 75 RDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLPLGKVPEGTVVCSVEEKAGDR 134
R+A ++ + +A EG+H GQF++CGKK L IG+VL + +PE T VC ++EK DR
Sbjct 54 RNAPKW---TQLFIAAEGIHMGQFVYCGKKVQLNIGSVLSVDTMPESTTVCCLKEKPEDR 110
Query 135 GTIAKTSGTFATVVGHSEETGKT 157
G +A+ SG ATV+ H+ ET KT
Sbjct 111 GKLARASGNGATVISHNPETQKT 133
> ECU01g0310
Length=239
Score = 88.6 bits (218), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 47/117 (40%), Positives = 72/117 (61%), Gaps = 6/117 (5%)
Query 53 KGLVTSIEHDPGRGAPLAVVSFRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNV 112
+G V I H+ RGAP+A++SF++ + + A EG++TGQ I G A + IGN+
Sbjct 34 EGEVIDIVHERSRGAPVAIISFKE------DDCMVPASEGMYTGQKILIGDDAPIDIGNI 87
Query 113 LPLGKVPEGTVVCSVEEKAGDRGTIAKTSGTFATVVGHSEETGKTRVRLPSGARKTL 169
+ VPEG V SVE GD GT A +G+++ VV H +ET +T +++PSG + T+
Sbjct 88 TKIKNVPEGMAVNSVESVYGDGGTFAMVNGSYSLVVNHRKETNETVIKIPSGKKLTV 144
> AtCh087
Length=274
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 47/170 (27%), Positives = 77/170 (45%), Gaps = 20/170 (11%)
Query 4 QQQQEDGTRHPLLLAQ------RKGRGSVFRAHT---HKRKGAAKLRALDYAERNGYIKG 54
Q + R+ L+ Q R RG + H HKR R +D+ I G
Sbjct 19 DSQVKSNPRNNLICGQHHCGKGRNARGIITARHRGGGHKRL----YRKIDFRRNAKDIYG 74
Query 55 LVTSIEHDPGRGAPLAVVSFRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLP 114
+ +IE+DP R A + ++ + D K +L G G I G + + +GN LP
Sbjct 75 RIVTIEYDPNRNAYICLIHYGDG-----EKRYILHPRGAIIGDTIVSGTEVPIKMGNALP 129
Query 115 LGKVPEGTVVCSVEEKAGDRGTIAKTSGTFATVVGHSEETGKTRVRLPSG 164
L +P GT + ++E G G +A+ +G A ++ ++E ++LPSG
Sbjct 130 LTDMPLGTAIHNIEITLGRGGQLARAAGAVAKLI--AKEGKSATLKLPSG 177
> AtCh063
Length=274
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 47/170 (27%), Positives = 77/170 (45%), Gaps = 20/170 (11%)
Query 4 QQQQEDGTRHPLLLAQ------RKGRGSVFRAHT---HKRKGAAKLRALDYAERNGYIKG 54
Q + R+ L+ Q R RG + H HKR R +D+ I G
Sbjct 19 DSQVKSNPRNNLICGQHHCGKGRNARGIITARHRGGGHKRL----YRKIDFRRNAKDIYG 74
Query 55 LVTSIEHDPGRGAPLAVVSFRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTIGNVLP 114
+ +IE+DP R A + ++ + D K +L G G I G + + +GN LP
Sbjct 75 RIVTIEYDPNRNAYICLIHYGDG-----EKRYILHPRGAIIGDTIVSGTEVPIKMGNALP 129
Query 115 LGKVPEGTVVCSVEEKAGDRGTIAKTSGTFATVVGHSEETGKTRVRLPSG 164
L +P GT + ++E G G +A+ +G A ++ ++E ++LPSG
Sbjct 130 LTDMPLGTAIHNIEITLGRGGQLARAAGAVAKLI--AKEGKSATLKLPSG 177
> Hs17459950
Length=124
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 22/44 (50%), Positives = 33/44 (75%), Gaps = 0/44 (0%)
Query 124 VCSVEEKAGDRGTIAKTSGTFATVVGHSEETGKTRVRLPSGARK 167
+C +EEK D G + + SG +ATV+ H+ ET K+RV+LPSG++K
Sbjct 1 MCCLEEKPRDHGKLTQASGNYATVISHNPETKKSRVKLPSGSKK 44
> 7294756
Length=294
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 38/120 (31%), Positives = 59/120 (49%), Gaps = 7/120 (5%)
Query 48 RNGYIKG----LVTSIEHDPGRGAPLAVVSFRDAYRYRINKERMLAVEGLHTGQFIHCGK 103
R+G +G LV + D R A +A+V+ D +Y + E M A + L T +FI
Sbjct 96 RDGPTEGAQEELVLEVLRDGCRTAKVALVAVGDELKYILATENMKAGDILKTSRFIP-RI 154
Query 104 KADLTIGNVLPLGKVPEGTVVCSVEEKAGDRGTIAKTSGTFATVVGHSEETGKTRVRLPS 163
G+ PLG +P GT + +E+ G + +GTF T++ +E K V+LPS
Sbjct 155 PVRPNEGDAYPLGALPVGTRIHCLEKNPGQMCHLIHAAGTFGTILRKFDE--KVVVQLPS 212
> YEL050c
Length=393
Score = 45.1 bits (105), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 39/147 (26%), Positives = 62/147 (42%), Gaps = 26/147 (17%)
Query 39 KLRALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSFRDAYRYRINKERMLAVEGLHTGQF 98
+ R +D+ G + V IE+DPGR + +A++ ++A +GL G
Sbjct 151 RTRLIDFNRWEGGAQ-TVQRIEYDPGRSSHIALLKHNTTGEL----SYIIACDGLRPGDV 205
Query 99 IHC-------------GKKADLTI--------GNVLPLGKVPEGTVVCSVEEKAGDRGTI 137
+ G K D I GN LP+ +P GT++ +V G
Sbjct 206 VESFRRGIPQTLLNEMGGKVDPAILSVKTTQRGNCLPISMIPIGTIIHNVGITPVGPGKF 265
Query 138 AKTSGTFATVVGHSEETGKTRVRLPSG 164
+++GT+A V+ E K VRL SG
Sbjct 266 CRSAGTYARVLAKLPEKKKAIVRLQSG 292
> HsM7705714
Length=353
Score = 41.6 bits (96), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 42/162 (25%), Positives = 73/162 (45%), Gaps = 13/162 (8%)
Query 11 TRHPLLLAQRKGRGSVFRAHTHKRKGAAK--LRALDYAE------RNGYIKGLVTSIEHD 62
T P+ + + GR R H G K R +D+ ++G + V + +D
Sbjct 73 TITPVKMRKSGGRDHTGRIRVHGIGGGHKQRYRMIDFLRFRPEETKSGPFEEKVIQVRYD 132
Query 63 PGRGAPLAVVSFRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLT-IGNVLPLGKVPEG 121
P R A +A+V+ R+ I E M A + + H G+ A G+ PLG +P G
Sbjct 133 PCRSADIALVAGGSRKRWIIATENMQAGDTILNSN--HIGRMAVAAREGDAHPLGALPVG 190
Query 122 TVVCSVEEKAGDRGTIAKTSGTFATVVGHSEETGKTRVRLPS 163
T++ +VE + G + +GT ++ + G + ++LPS
Sbjct 191 TLINNVESEPGRGAQYIRAAGTCGVLL--RKVNGHSIIQLPS 230
> Hs21265070
Length=305
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 41/159 (25%), Positives = 71/159 (44%), Gaps = 13/159 (8%)
Query 14 PLLLAQRKGRGSVFRAHTHKRKGAAK--LRALDYAE------RNGYIKGLVTSIEHDPGR 65
P+ + + GR R H G K R +D+ ++G + V + +DP R
Sbjct 76 PVKMRKSGGRDHTGRIRVHGIGGGHKQRYRMIDFLRFRPEETKSGPFEEKVIQVRYDPCR 135
Query 66 GAPLAVVSFRDAYRYRINKERMLAVEGLHTGQFIHCGKKADLTI-GNVLPLGKVPEGTVV 124
A +A+V+ R+ I E M A + + H G+ A G+ PLG +P GT++
Sbjct 136 SADIALVAGGSRKRWIIATENMQAGDTILNSN--HIGRMAVAAREGDAHPLGALPVGTLI 193
Query 125 CSVEEKAGDRGTIAKTSGTFATVVGHSEETGKTRVRLPS 163
+VE + G + +GT ++ + G ++LPS
Sbjct 194 NNVESEPGRGAQYIRAAGTCGVLL--RKVNGTAIIQLPS 230
> SPCC16C4.15
Length=318
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 45/190 (23%), Positives = 79/190 (41%), Gaps = 41/190 (21%)
Query 14 PLLLAQRK--GRGSVFRAHTHKRKGAAK--LRALDYAERNGYIKGL--VTSIEHDPGRGA 67
PL +A+RK GR R + G K +R +D+ + + G+ V IE+DPGR
Sbjct 40 PLTVAKRKKGGRNETGRITVRHQGGGHKQRIRLVDFERK---VPGVHRVIRIEYDPGRSG 96
Query 68 PLAVVSFRDAYRYRINKERMLAVEGLHTGQFIHCGKK----------------------- 104
+A+V ++ NK +LA +GL G + +
Sbjct 97 HIALVEKLNS--ETANKSYILACDGLREGDTVESFRSLLKTGSNGEPVEMDPVLAAEIED 154
Query 105 -----ADLTIGNVLPLGKVPEGTVVCSVEEKAGDRGTIAKTSGTFATVVGHSEETGKTRV 159
+L GN PL +P GTV+ ++ + + +++G+ A ++ + V
Sbjct 155 GTFAAKNLKPGNCFPLRLIPIGTVIHAIGVNPNQKAKLCRSAGSSARIIAFDGKYAI--V 212
Query 160 RLPSGARKTL 169
RL SG + +
Sbjct 213 RLQSGEERKI 222
> CE18483_2
Length=72
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 15/28 (53%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 142 GTFATVVGHSEETGKTRVRLPSGARKTL 169
G +A V+ H+ +T KTR+RLPSGA+K +
Sbjct 1 GNYANVIAHNADTKKTRIRLPSGAKKVV 28
> AtMi044
Length=349
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 30/55 (54%), Gaps = 3/55 (5%)
Query 22 GRGSVFRAHTHKRKGAAK--LRALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSF 74
GR S R R G +K LR +D +R+ G+V SIE+DP R + +A V +
Sbjct 21 GRNSSGRITVFHRGGGSKRLLRRIDL-KRSTSSMGIVESIEYDPNRSSQIAPVRW 74
> At2g07715
Length=307
Score = 30.0 bits (66), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 30/55 (54%), Gaps = 3/55 (5%)
Query 22 GRGSVFRAHTHKRKGAAK--LRALDYAERNGYIKGLVTSIEHDPGRGAPLAVVSF 74
GR S R R G +K LR +D +R+ G+V SIE+DP R + +A V +
Sbjct 21 GRNSSGRITVFHRGGGSKRLLRRIDL-KRSTSSMGIVESIEYDPNRSSQIAPVRW 74
> Hs22061364
Length=197
Score = 29.6 bits (65), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 31/54 (57%), Gaps = 2/54 (3%)
Query 110 GNVLPLGKVPEGTVVCSVEEKAGDRGTIAKTSGTFATVVGHSEETGKTRVRLPS 163
G+ PLG +P GT++ ++E + G + +GT + ++ + G T ++LPS
Sbjct 84 GDAHPLGALPVGTLINNMEIEPGRGAQYIQAAGTCSVLLW--KVNGTTVIQLPS 135
> At4g30170
Length=325
Score = 28.5 bits (62), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 11/27 (40%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 79 RYRINKERMLAVEGLHTGQFIHCGKKA 105
R+ +++ M+A+ G HT F HCGK +
Sbjct 178 RHGLSQTDMIALSGAHTIGFAHCGKMS 204
> At5g67400
Length=329
Score = 28.5 bits (62), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 10/25 (40%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 79 RYRINKERMLAVEGLHTGQFIHCGK 103
+ ++ +E M+A+ HT F HCGK
Sbjct 182 KNKLTQEDMIALSAAHTLGFAHCGK 206
> At2g18980
Length=323
Score = 28.1 bits (61), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 11/25 (44%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 79 RYRINKERMLAVEGLHTGQFIHCGK 103
R+ +++ M+A+ G HT F HCGK
Sbjct 176 RHGLSQTDMIALSGAHTIGFAHCGK 200
> Hs20484677
Length=293
Score = 28.1 bits (61), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 10/27 (37%), Positives = 20/27 (74%), Gaps = 0/27 (0%)
Query 29 AHTHKRKGAAKLRALDYAERNGYIKGL 55
AH +K+ G+ +++A + +E NG+I G+
Sbjct 33 AHIYKKHGSQRVKARELSEHNGHITGI 59
Lambda K H
0.317 0.134 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2490594054
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40