bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_2593_orf3
Length=293
Score E
Sequences producing significant alignments: (Bits) Value
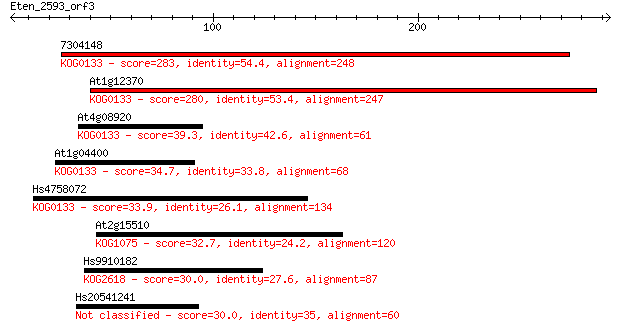
7304148 283 3e-76
At1g12370 280 2e-75
At4g08920 39.3 0.010
At1g04400 34.7 0.21
Hs4758072 33.9 0.42
At2g15510 32.7 0.97
Hs9910182 30.0 5.3
Hs20541241 30.0 5.8
> 7304148
Length=555
Score = 283 bits (723), Expect = 3e-76, Method: Compositional matrix adjust.
Identities = 135/248 (54%), Positives = 167/248 (67%), Gaps = 6/248 (2%)
Query 26 LKADNRVQLRHCWRPGATAGLKAMKVFCTVPKLLAYTSRNNPVLDSQSGLSPWLHFGQIS 85
L+ D V +PG A + + FC+ RN+P D+ SGLSPWLHFG IS
Sbjct 303 LQCDMEVDEVQWAKPGYKAACQQLYEFCSRRLRHFNDKRNDPTADALSGLSPWLHFGHIS 362
Query 86 AQRCLASVKDMGPATKIGATTAAARESYIEELVVRRELAENFTFYNSKYNQITGAPIWAQ 145
AQRC V+ + A+ +++ EE +VRRELA+NF FYN Y+ + G WA
Sbjct 363 AQRCALEVQ------RFRGQHKASADAFCEEAIVRRELADNFCFYNEHYDSLKGLSSWAY 416
Query 146 ETLAAHKNDKREHLYALEQLEGSKTHDELWNAAQLQLVHTAKMHGFLRMYWAKKILEWTP 205
+TL AH+ DKR+ Y+LE+LE S T+D+LWN+AQLQLV KMHGFLRMYWAKKILEWT
Sbjct 417 QTLDAHRKDKRDPCYSLEELEKSLTYDDLWNSAQLQLVREGKMHGFLRMYWAKKILEWTA 476
Query 206 SADIALTTALFLNDKFSLDGTDPNGVVGCMWSVAGVHDQGWAERPIFGKIRYMNLAGCMR 265
+ + AL A+ LNDK+SLDG DPNG VGCMWS+ GVHD GW ER IFGK+RYMN GC R
Sbjct 477 TPEHALEYAILLNDKYSLDGRDPNGYVGCMWSIGGVHDMGWKERAIFGKVRYMNYQGCRR 536
Query 266 KFSVDAFV 273
KF V+AFV
Sbjct 537 KFDVNAFV 544
> At1g12370
Length=496
Score = 280 bits (717), Expect = 2e-75, Method: Compositional matrix adjust.
Identities = 132/252 (52%), Positives = 181/252 (71%), Gaps = 11/252 (4%)
Query 40 PGATAGLKAM---KVFCTVPKLLAY-TSRNNPVL-DSQSGLSPWLHFGQISAQRCLASVK 94
PG AG++ + K +L Y T RNNP+ + SGLSP+LHFGQ+SAQRC
Sbjct 232 PGEDAGIEVLMGNKDGFLTKRLKNYSTDRNNPIKPKALSGLSPYLHFGQVSAQRCALE-- 289
Query 95 DMGPATKIGATTAAARESYIEELVVRRELAENFTFYNSKYNQITGAPIWAQETLAAHKND 154
A K+ +T+ A ++++EEL+VRREL++NF +Y Y+ + GA WA+++L H +D
Sbjct 290 ----ARKVRSTSPQAVDTFLEELIVRRELSDNFCYYQPHYDSLKGAWEWARKSLMDHASD 345
Query 155 KREHLYALEQLEGSKTHDELWNAAQLQLVHTAKMHGFLRMYWAKKILEWTPSADIALTTA 214
KREH+Y+LEQLE T D LWNA+QL++V+ KMHGF+RMYWAKKILEWT + AL+ +
Sbjct 346 KREHIYSLEQLEKGLTADPLWNASQLEMVYQGKMHGFMRMYWAKKILEWTKGPEEALSIS 405
Query 215 LFLNDKFSLDGTDPNGVVGCMWSVAGVHDQGWAERPIFGKIRYMNLAGCMRKFSVDAFVR 274
++LN+K+ +DG DP+G VGCMWS+ GVHDQGW ERP+FGKIRYMN AGC RKF+VD+++
Sbjct 406 IYLNNKYEIDGRDPSGYVGCMWSICGVHDQGWKERPVFGKIRYMNYAGCKRKFNVDSYIS 465
Query 275 CVRQIVGRHAKK 286
V+ +V KK
Sbjct 466 YVKSLVSVTKKK 477
> At4g08920
Length=716
Score = 39.3 bits (90), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 26/63 (41%), Positives = 34/63 (53%), Gaps = 4/63 (6%)
Query 34 LRHCWRPGATAGLKAMKVFCTVPKLLAYTSRNNPVLDS--QSGLSPWLHFGQISAQRCLA 91
L W PG + G KA+ F P LL Y S+N DS S LSP LHFG++S ++
Sbjct 209 LARAWSPGWSNGDKALTTFINGP-LLEY-SKNRRKADSATTSFLSPHLHFGEVSVRKVFH 266
Query 92 SVK 94
V+
Sbjct 267 LVR 269
> At1g04400
Length=612
Score = 34.7 bits (78), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 37/75 (49%), Gaps = 8/75 (10%)
Query 23 LEQLKADNRVQ------LRHCWRPGATAGLKAMKVFCTVPKLLAYTSRNNPVL-DSQSGL 75
+E+L +N + L W PG + K + F +L+ Y + V+ +S S L
Sbjct 189 IEELGLENEAEKPSNALLTRAWSPGWSNADKLLNEFIE-KQLIDYAKNSKKVVGNSTSLL 247
Query 76 SPWLHFGQISAQRCL 90
SP+LHFG+IS +
Sbjct 248 SPYLHFGEISVRHVF 262
> Hs4758072
Length=586
Score = 33.9 bits (76), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 35/138 (25%), Positives = 57/138 (41%), Gaps = 10/138 (7%)
Query 12 HGEDARFPSDILEQLKADNRVQLRHCWRPGATAGLKAMKVFCTVPKLLAYTSRN----NP 67
H E PS LE+L D W G T L ++ +A R N
Sbjct 186 HDEKYGVPS--LEELGFDTDGLSSAVWPGGETEALTRLERHLERKAWVANFERPRMNANS 243
Query 68 VLDSQSGLSPWLHFGQISAQRCLASVKDMGPATKIGATTAAARESYIEELVVRRELAENF 127
+L S +GLSP+L FG +S + + D+ K+ ++ Y + ++ RE
Sbjct 244 LLASPTGLSPYLRFGCLSCRLFYFKLTDL--YKKVKKNSSPPLSLYGQ--LLWREFFYTA 299
Query 128 TFYNSKYNQITGAPIWAQ 145
N +++++ G PI Q
Sbjct 300 ATNNPRFDKMEGNPICVQ 317
> At2g15510
Length=1138
Score = 32.7 bits (73), Expect = 0.97, Method: Compositional matrix adjust.
Identities = 29/123 (23%), Positives = 51/123 (41%), Gaps = 11/123 (8%)
Query 43 TAGLKAMKVFCTVPKLLAYTSRNNPVLDSQSGLSPWLHFGQISAQRCLASVKDMGPATKI 102
++G KV T+ L +++ SQ + W+H G + ASVKD
Sbjct 275 SSGFPRSKVVFTLKNDLCKANQDEETFWSQKSRAKWMHGGDKNTSFFHASVKDNRDEVSK 334
Query 103 GATTAAARESYIEELVVRRE---LAENFTFYNSKYNQITGAPIWAQETLAAHKNDKREHL 159
GA ESY ++L E A+ F + + ++ + T KN+ R+ +
Sbjct 335 GAIA----ESYFKDLFKSSEGSNFADLFAGFQPRVTEVMNRVL----TATVSKNEVRDAV 386
Query 160 YAL 162
+A+
Sbjct 387 FAI 389
> Hs9910182
Length=371
Score = 30.0 bits (66), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 24/97 (24%), Positives = 39/97 (40%), Gaps = 10/97 (10%)
Query 37 CWRPGATAGLKAMKVFCTVPKLLAYTS---RNNPVLDSQSGLSPWLHFGQISAQRCLASV 93
CW P + TV + R +LD + +S WL FG++S Q L +
Sbjct 182 CWEPPEGGDVLYSYTIITVDSCKGLSDIHHRMPAILDGEEAVSKWLDFGEVSTQEALKLI 241
Query 94 KDMGPAT--KIGATTAAARESYIE-----ELVVRREL 123
T + + +R + E +LVV++EL
Sbjct 242 HPTENITFHAVSSVVNNSRNNTPECLAPVDLVVKKEL 278
> Hs20541241
Length=265
Score = 30.0 bits (66), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 30/60 (50%), Gaps = 1/60 (1%)
Query 33 QLRHCWRPGATAGLKAMKVFCTVPKLLAYTSRNNPVLDSQSGLSPWLHFGQISAQRCLAS 92
Q HC+R G KA++V C V KLL R +++ + L+ W I+ CLAS
Sbjct 154 QKSHCFRLGRQLS-KALQVNCVVRKLLVQLRRLYWWVETMTALTSWHLAYLITWTTCLAS 212
Lambda K H
0.320 0.133 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6531448310
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40